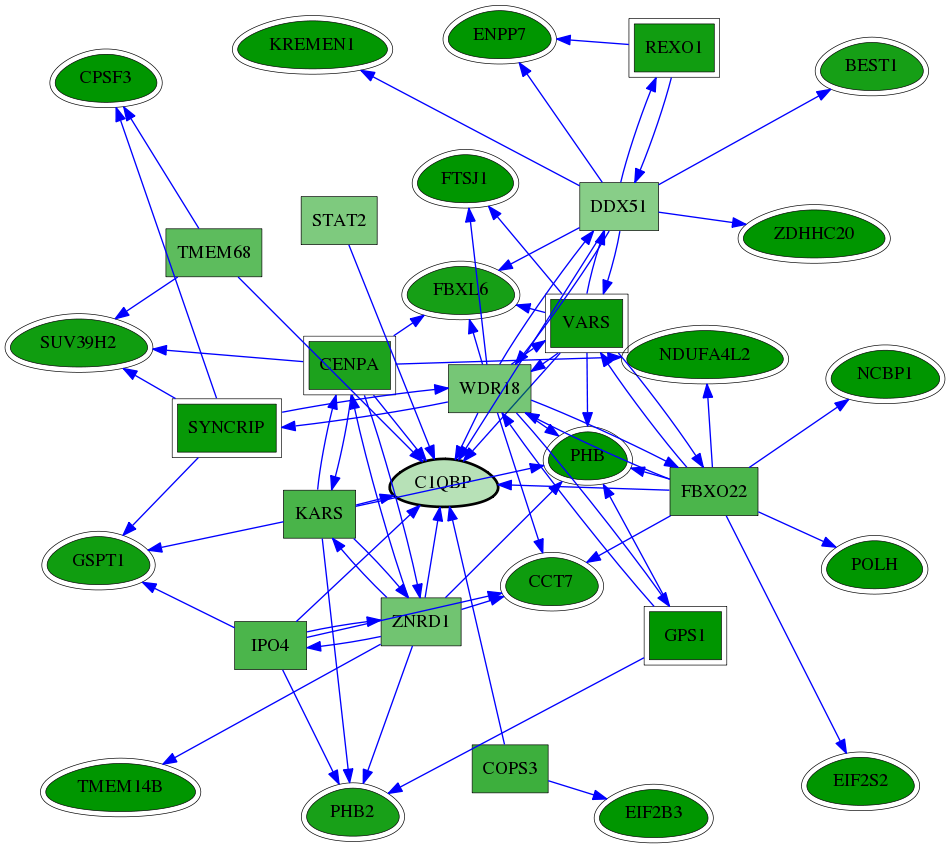
| GO:0010388 | cullin deneddylation | 3.06e-04 | 1.00e+00 | 6.209 | 2 | 4 | 8 |
| GO:0006379 | mRNA cleavage | 3.93e-04 | 1.00e+00 | 6.039 | 2 | 2 | 9 |
| GO:0008135 | translation factor activity, nucleic acid binding | 1.13e-03 | 1.00e+00 | 5.302 | 2 | 4 | 15 |
| GO:0008380 | RNA splicing | 2.19e-03 | 1.00e+00 | 2.843 | 4 | 21 | 165 |
| GO:0030529 | ribonucleoprotein complex | 2.60e-03 | 1.00e+00 | 3.436 | 3 | 5 | 82 |
| GO:0016032 | viral process | 2.77e-03 | 1.00e+00 | 2.053 | 6 | 30 | 428 |
| GO:0008180 | COP9 signalosome | 2.92e-03 | 1.00e+00 | 4.624 | 2 | 6 | 24 |
| GO:0004832 | valine-tRNA ligase activity | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0006430 | lysyl-tRNA aminoacylation | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0002128 | tRNA nucleoside ribose methylation | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0008175 | tRNA methyltransferase activity | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0097177 | mitochondrial ribosome binding | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0000939 | condensed chromosome inner kinetochore | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0030984 | kininogen binding | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0006438 | valyl-tRNA aminoacylation | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0004824 | lysine-tRNA ligase activity | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0031690 | adrenergic receptor binding | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 1 |
| GO:0006418 | tRNA aminoacylation for protein translation | 4.25e-03 | 1.00e+00 | 4.351 | 2 | 4 | 29 |
| GO:0005739 | mitochondrion | 5.63e-03 | 1.00e+00 | 1.652 | 7 | 32 | 659 |
| GO:0031124 | mRNA 3'-end processing | 6.15e-03 | 1.00e+00 | 4.080 | 2 | 4 | 35 |
| GO:0039536 | negative regulation of RIG-I signaling pathway | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0051716 | cellular response to stimulus | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:1901165 | positive regulation of trophoblast cell migration | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0006684 | sphingomyelin metabolic process | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0005846 | nuclear cap binding complex | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0002176 | male germ cell proliferation | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0039534 | negative regulation of MDA-5 signaling pathway | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0048742 | regulation of skeletal muscle fiber development | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0003747 | translation release factor activity | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 2 | 2 |
| GO:0015966 | diadenosine tetraphosphate biosynthetic process | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0070131 | positive regulation of mitochondrial translation | 6.75e-03 | 1.00e+00 | 7.209 | 1 | 1 | 2 |
| GO:0003743 | translation initiation factor activity | 6.85e-03 | 1.00e+00 | 4.000 | 2 | 8 | 37 |
| GO:0006369 | termination of RNA polymerase II transcription | 7.22e-03 | 1.00e+00 | 3.961 | 2 | 5 | 38 |
| GO:0005654 | nucleoplasm | 7.42e-03 | 1.00e+00 | 1.434 | 8 | 68 | 876 |
| GO:0010467 | gene expression | 8.21e-03 | 1.00e+00 | 1.731 | 6 | 49 | 535 |
| GO:0000775 | chromosome, centromeric region | 8.36e-03 | 1.00e+00 | 3.851 | 2 | 6 | 41 |
| GO:0000398 | mRNA splicing, via spliceosome | 9.01e-03 | 1.00e+00 | 2.794 | 3 | 15 | 128 |
| GO:0003723 | RNA binding | 9.19e-03 | 1.00e+00 | 2.261 | 4 | 22 | 247 |
| GO:0006406 | mRNA export from nucleus | 9.59e-03 | 1.00e+00 | 3.750 | 2 | 7 | 44 |
| GO:0036123 | histone H3-K9 dimethylation | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 1 | 3 |
| GO:0000778 | condensed nuclear chromosome kinetochore | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 2 | 3 |
| GO:0042256 | mature ribosome assembly | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 1 | 3 |
| GO:0071459 | protein localization to chromosome, centromeric region | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 1 | 3 |
| GO:0004767 | sphingomyelin phosphodiesterase activity | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 1 | 3 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 2 | 3 |
| GO:2000510 | positive regulation of dendritic cell chemotaxis | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 1 | 3 |
| GO:0060828 | regulation of canonical Wnt signaling pathway | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0097452 | GAIT complex | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 2 | 4 |
| GO:0030321 | transepithelial chloride transport | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 2 | 4 |
| GO:0045292 | mRNA cis splicing, via spliceosome | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0031442 | positive regulation of mRNA 3'-end processing | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0006290 | pyrimidine dimer repair | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 2 | 4 |
| GO:0032057 | negative regulation of translational initiation in response to stress | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0036124 | histone H3-K9 trimethylation | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 2 | 4 |
| GO:0006333 | chromatin assembly or disassembly | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0016779 | nucleotidyltransferase activity | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 1.35e-02 | 1.00e+00 | 6.209 | 1 | 1 | 4 |
| GO:0016020 | membrane | 1.59e-02 | 1.00e+00 | 1.142 | 9 | 56 | 1207 |
| GO:0070934 | CRD-mediated mRNA stabilization | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 2 | 5 |
| GO:0001849 | complement component C1q binding | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 2 | 5 |
| GO:0060744 | mammary gland branching involved in thelarche | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:0002161 | aminoacyl-tRNA editing activity | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:0004521 | endoribonuclease activity | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:0006398 | histone mRNA 3'-end processing | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:0006282 | regulation of DNA repair | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 2 | 5 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 1 | 5 |
| GO:0050847 | progesterone receptor signaling pathway | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 2 | 6 |
| GO:0002181 | cytoplasmic translation | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 1 | 6 |
| GO:0042754 | negative regulation of circadian rhythm | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 2 | 6 |
| GO:0046974 | histone methyltransferase activity (H3-K9 specific) | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 2 | 6 |
| GO:0003676 | nucleic acid binding | 2.08e-02 | 1.00e+00 | 3.165 | 2 | 4 | 66 |
| GO:0002199 | zona pellucida receptor complex | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 4 | 7 |
| GO:0007140 | male meiosis | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 1 | 7 |
| GO:0008409 | 5'-3' exonuclease activity | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 2 | 7 |
| GO:0000339 | RNA cap binding | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 1 | 7 |
| GO:0050687 | negative regulation of defense response to virus | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 1 | 7 |
| GO:0051382 | kinetochore assembly | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 2 | 7 |
| GO:0000731 | DNA synthesis involved in DNA repair | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 1 | 7 |
| GO:0006301 | postreplication repair | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 2 | 7 |
| GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 2 | 7 |
| GO:0032695 | negative regulation of interleukin-12 production | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 1 | 7 |
| GO:0000780 | condensed nuclear chromosome, centromeric region | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 1 | 7 |
| GO:0005832 | chaperonin-containing T-complex | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 4 | 7 |
| GO:0001649 | osteoblast differentiation | 2.57e-02 | 1.00e+00 | 3.000 | 2 | 5 | 74 |
| GO:0005845 | mRNA cap binding complex | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 1 | 8 |
| GO:0006450 | regulation of translational fidelity | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 1 | 8 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 1 | 8 |
| GO:0071354 | cellular response to interleukin-6 | 3.00e-02 | 1.00e+00 | 5.039 | 1 | 1 | 9 |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 3.00e-02 | 1.00e+00 | 5.039 | 1 | 3 | 9 |
| GO:0050908 | detection of light stimulus involved in visual perception | 3.00e-02 | 1.00e+00 | 5.039 | 1 | 1 | 9 |
| GO:0010225 | response to UV-C | 3.00e-02 | 1.00e+00 | 5.039 | 1 | 2 | 9 |
| GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 3.00e-02 | 1.00e+00 | 5.039 | 1 | 2 | 9 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 3.25e-02 | 1.00e+00 | 2.817 | 2 | 5 | 84 |
| GO:0009416 | response to light stimulus | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 1 | 10 |
| GO:0001054 | RNA polymerase I activity | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 4 | 10 |
| GO:0005736 | DNA-directed RNA polymerase I complex | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 4 | 10 |
| GO:0005095 | GTPase inhibitor activity | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 1 | 10 |
| GO:0008143 | poly(A) binding | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 2 | 10 |
| GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 1 | 10 |
| GO:0008334 | histone mRNA metabolic process | 3.66e-02 | 1.00e+00 | 4.750 | 1 | 1 | 11 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 3.66e-02 | 1.00e+00 | 4.750 | 1 | 4 | 11 |
| GO:0006479 | protein methylation | 3.66e-02 | 1.00e+00 | 4.750 | 1 | 2 | 11 |
| GO:0005540 | hyaluronic acid binding | 3.66e-02 | 1.00e+00 | 4.750 | 1 | 1 | 11 |
| GO:0007154 | cell communication | 3.66e-02 | 1.00e+00 | 4.750 | 1 | 1 | 11 |
| GO:0004527 | exonuclease activity | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 4 | 12 |
| GO:0000132 | establishment of mitotic spindle orientation | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 2 | 12 |
| GO:0016409 | palmitoyltransferase activity | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 1 | 12 |
| GO:0006336 | DNA replication-independent nucleosome assembly | 4.31e-02 | 1.00e+00 | 4.509 | 1 | 2 | 13 |
| GO:0007597 | blood coagulation, intrinsic pathway | 4.31e-02 | 1.00e+00 | 4.509 | 1 | 2 | 13 |
| GO:0008156 | negative regulation of DNA replication | 4.31e-02 | 1.00e+00 | 4.509 | 1 | 1 | 13 |
| GO:0006335 | DNA replication-dependent nucleosome assembly | 4.31e-02 | 1.00e+00 | 4.509 | 1 | 2 | 13 |
| GO:0060749 | mammary gland alveolus development | 4.31e-02 | 1.00e+00 | 4.509 | 1 | 1 | 13 |
| GO:0006413 | translational initiation | 4.38e-02 | 1.00e+00 | 2.580 | 2 | 6 | 99 |
| GO:0016746 | transferase activity, transferring acyl groups | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 2 | 14 |
| GO:0090023 | positive regulation of neutrophil chemotaxis | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 1 | 14 |
| GO:0030449 | regulation of complement activation | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 2 | 14 |
| GO:0016597 | amino acid binding | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 1 | 14 |
| GO:0016575 | histone deacetylation | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 2 | 14 |
| GO:0051924 | regulation of calcium ion transport | 4.95e-02 | 1.00e+00 | 4.302 | 1 | 1 | 15 |
| GO:0000188 | inactivation of MAPK activity | 4.95e-02 | 1.00e+00 | 4.302 | 1 | 1 | 15 |
| GO:0018345 | protein palmitoylation | 4.95e-02 | 1.00e+00 | 4.302 | 1 | 1 | 15 |
| GO:0014003 | oligodendrocyte development | 4.95e-02 | 1.00e+00 | 4.302 | 1 | 1 | 15 |
| GO:0007339 | binding of sperm to zona pellucida | 4.95e-02 | 1.00e+00 | 4.302 | 1 | 4 | 15 |
| GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 1 | 16 |
| GO:0014065 | phosphatidylinositol 3-kinase signaling | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 1 | 16 |
| GO:0060338 | regulation of type I interferon-mediated signaling pathway | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 1 | 16 |
| GO:0003887 | DNA-directed DNA polymerase activity | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 2 | 16 |
| GO:0005515 | protein binding | 5.45e-02 | 1.00e+00 | 0.447 | 19 | 198 | 4124 |
| GO:0032689 | negative regulation of interferon-gamma production | 5.60e-02 | 1.00e+00 | 4.122 | 1 | 1 | 17 |
| GO:0019706 | protein-cysteine S-palmitoyltransferase activity | 5.60e-02 | 1.00e+00 | 4.122 | 1 | 1 | 17 |
| GO:0031047 | gene silencing by RNA | 5.92e-02 | 1.00e+00 | 4.039 | 1 | 2 | 18 |
| GO:0006378 | mRNA polyadenylation | 5.92e-02 | 1.00e+00 | 4.039 | 1 | 3 | 18 |
| GO:0005829 | cytosol | 6.54e-02 | 1.00e+00 | 0.728 | 10 | 88 | 1787 |
| GO:0006913 | nucleocytoplasmic transport | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 2 | 20 |
| GO:0071346 | cellular response to interferon-gamma | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 2 | 20 |
| GO:0005720 | nuclear heterochromatin | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 3 | 20 |
| GO:0005254 | chloride channel activity | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 1 | 20 |
| GO:0034660 | ncRNA metabolic process | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 1 | 20 |
| GO:0034080 | CENP-A containing nucleosome assembly | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 2 | 20 |
| GO:0071897 | DNA biosynthetic process | 6.55e-02 | 1.00e+00 | 3.887 | 1 | 2 | 20 |
| GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 6.87e-02 | 1.00e+00 | 3.817 | 1 | 2 | 21 |
| GO:0008536 | Ran GTPase binding | 6.87e-02 | 1.00e+00 | 3.817 | 1 | 1 | 21 |
| GO:0006363 | termination of RNA polymerase I transcription | 6.87e-02 | 1.00e+00 | 3.817 | 1 | 3 | 21 |
| GO:0006139 | nucleobase-containing compound metabolic process | 7.18e-02 | 1.00e+00 | 3.750 | 1 | 2 | 22 |
| GO:0051084 | 'de novo' posttranslational protein folding | 7.18e-02 | 1.00e+00 | 3.750 | 1 | 4 | 22 |
| GO:0030331 | estrogen receptor binding | 7.18e-02 | 1.00e+00 | 3.750 | 1 | 2 | 22 |
| GO:0000387 | spliceosomal snRNP assembly | 7.18e-02 | 1.00e+00 | 3.750 | 1 | 1 | 22 |
| GO:0006687 | glycosphingolipid metabolic process | 7.18e-02 | 1.00e+00 | 3.750 | 1 | 1 | 22 |
| GO:0006821 | chloride transport | 7.50e-02 | 1.00e+00 | 3.685 | 1 | 2 | 23 |
| GO:0000049 | tRNA binding | 7.50e-02 | 1.00e+00 | 3.685 | 1 | 2 | 23 |
| GO:0006370 | 7-methylguanosine mRNA capping | 7.81e-02 | 1.00e+00 | 3.624 | 1 | 5 | 24 |
| GO:0034707 | chloride channel complex | 7.81e-02 | 1.00e+00 | 3.624 | 1 | 1 | 24 |
| GO:0005759 | mitochondrial matrix | 7.98e-02 | 1.00e+00 | 2.090 | 2 | 7 | 139 |
| GO:0005902 | microvillus | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 1 | 25 |
| GO:0008033 | tRNA processing | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 2 | 25 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 3 | 25 |
| GO:0007259 | JAK-STAT cascade | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 2 | 25 |
| GO:0009267 | cellular response to starvation | 8.44e-02 | 1.00e+00 | 3.509 | 1 | 1 | 26 |
| GO:0006958 | complement activation, classical pathway | 8.75e-02 | 1.00e+00 | 3.454 | 1 | 3 | 27 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 9.05e-02 | 1.00e+00 | 1.486 | 3 | 17 | 317 |
| GO:0045785 | positive regulation of cell adhesion | 9.36e-02 | 1.00e+00 | 3.351 | 1 | 3 | 29 |
| GO:0008026 | ATP-dependent helicase activity | 9.36e-02 | 1.00e+00 | 3.351 | 1 | 3 | 29 |
| GO:0006446 | regulation of translational initiation | 9.36e-02 | 1.00e+00 | 3.351 | 1 | 2 | 29 |
| GO:0001701 | in utero embryonic development | 9.50e-02 | 1.00e+00 | 1.942 | 2 | 7 | 154 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 9.98e-02 | 1.00e+00 | 3.255 | 1 | 6 | 31 |
| GO:0009408 | response to heat | 9.98e-02 | 1.00e+00 | 3.255 | 1 | 1 | 31 |
| GO:0005080 | protein kinase C binding | 9.98e-02 | 1.00e+00 | 3.255 | 1 | 2 | 31 |
| GO:0017148 | negative regulation of translation | 1.06e-01 | 1.00e+00 | 3.165 | 1 | 5 | 33 |
| GO:0000786 | nucleosome | 1.09e-01 | 1.00e+00 | 3.122 | 1 | 2 | 34 |
| GO:0007254 | JNK cascade | 1.09e-01 | 1.00e+00 | 3.122 | 1 | 1 | 34 |
| GO:0044297 | cell body | 1.12e-01 | 1.00e+00 | 3.080 | 1 | 5 | 35 |
| GO:0050434 | positive regulation of viral transcription | 1.12e-01 | 1.00e+00 | 3.080 | 1 | 5 | 35 |
| GO:0044267 | cellular protein metabolic process | 1.15e-01 | 1.00e+00 | 1.335 | 3 | 14 | 352 |
| GO:0043434 | response to peptide hormone | 1.15e-01 | 1.00e+00 | 3.039 | 1 | 1 | 36 |
| GO:1902476 | chloride transmembrane transport | 1.15e-01 | 1.00e+00 | 3.039 | 1 | 2 | 36 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1.21e-01 | 1.00e+00 | 2.961 | 1 | 4 | 38 |
| GO:0006665 | sphingolipid metabolic process | 1.21e-01 | 1.00e+00 | 2.961 | 1 | 1 | 38 |
| GO:0006412 | translation | 1.21e-01 | 1.00e+00 | 1.733 | 2 | 12 | 178 |
| GO:0048511 | rhythmic process | 1.21e-01 | 1.00e+00 | 2.961 | 1 | 5 | 38 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.22e-01 | 1.00e+00 | 1.725 | 2 | 17 | 179 |
| GO:0006396 | RNA processing | 1.27e-01 | 1.00e+00 | 2.887 | 1 | 4 | 40 |
| GO:0044822 | poly(A) RNA binding | 1.28e-01 | 1.00e+00 | 0.889 | 5 | 45 | 799 |
| GO:0003684 | damaged DNA binding | 1.30e-01 | 1.00e+00 | 2.851 | 1 | 1 | 41 |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 1.36e-01 | 1.00e+00 | 2.783 | 1 | 2 | 43 |
| GO:0060337 | type I interferon signaling pathway | 1.42e-01 | 1.00e+00 | 2.717 | 1 | 1 | 45 |
| GO:0009749 | response to glucose | 1.42e-01 | 1.00e+00 | 2.717 | 1 | 1 | 45 |
| GO:0005743 | mitochondrial inner membrane | 1.43e-01 | 1.00e+00 | 1.587 | 2 | 9 | 197 |
| GO:0005737 | cytoplasm | 1.53e-01 | 1.00e+00 | 0.431 | 12 | 127 | 2633 |
| GO:0043234 | protein complex | 1.58e-01 | 1.00e+00 | 1.495 | 2 | 13 | 210 |
| GO:0006334 | nucleosome assembly | 1.59e-01 | 1.00e+00 | 2.537 | 1 | 4 | 51 |
| GO:0005643 | nuclear pore | 1.59e-01 | 1.00e+00 | 2.537 | 1 | 12 | 51 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 1.62e-01 | 1.00e+00 | 2.509 | 1 | 8 | 52 |
| GO:0006338 | chromatin remodeling | 1.70e-01 | 1.00e+00 | 2.428 | 1 | 7 | 55 |
| GO:0008584 | male gonad development | 1.76e-01 | 1.00e+00 | 2.376 | 1 | 1 | 57 |
| GO:0003729 | mRNA binding | 1.76e-01 | 1.00e+00 | 2.376 | 1 | 4 | 57 |
| GO:0042826 | histone deacetylase binding | 1.79e-01 | 1.00e+00 | 2.351 | 1 | 5 | 58 |
| GO:0000785 | chromatin | 1.82e-01 | 1.00e+00 | 2.326 | 1 | 6 | 59 |
| GO:0071013 | catalytic step 2 spliceosome | 1.87e-01 | 1.00e+00 | 2.278 | 1 | 5 | 61 |
| GO:0006464 | cellular protein modification process | 1.87e-01 | 1.00e+00 | 2.278 | 1 | 4 | 61 |
| GO:0051897 | positive regulation of protein kinase B signaling | 1.90e-01 | 1.00e+00 | 2.255 | 1 | 1 | 62 |
| GO:0006415 | translational termination | 1.93e-01 | 1.00e+00 | 2.232 | 1 | 2 | 63 |
| GO:0030018 | Z disc | 1.95e-01 | 1.00e+00 | 2.209 | 1 | 3 | 64 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 1.98e-01 | 1.00e+00 | 0.961 | 3 | 29 | 456 |
| GO:0006364 | rRNA processing | 2.04e-01 | 1.00e+00 | 2.143 | 1 | 2 | 67 |
| GO:0003682 | chromatin binding | 2.09e-01 | 1.00e+00 | 1.232 | 2 | 15 | 252 |
| GO:0051082 | unfolded protein binding | 2.12e-01 | 1.00e+00 | 2.080 | 1 | 5 | 70 |
| GO:0030308 | negative regulation of cell growth | 2.12e-01 | 1.00e+00 | 2.080 | 1 | 3 | 70 |
| GO:0016363 | nuclear matrix | 2.20e-01 | 1.00e+00 | 2.019 | 1 | 10 | 73 |
| GO:0015630 | microtubule cytoskeleton | 2.25e-01 | 1.00e+00 | 1.980 | 1 | 5 | 75 |
| GO:0071456 | cellular response to hypoxia | 2.28e-01 | 1.00e+00 | 1.961 | 1 | 2 | 76 |
| GO:0008285 | negative regulation of cell proliferation | 2.40e-01 | 1.00e+00 | 1.095 | 2 | 11 | 277 |
| GO:0016055 | Wnt signaling pathway | 2.56e-01 | 1.00e+00 | 1.766 | 1 | 6 | 87 |
| GO:0006886 | intracellular protein transport | 2.61e-01 | 1.00e+00 | 1.733 | 1 | 2 | 89 |
| GO:0000209 | protein polyubiquitination | 2.69e-01 | 1.00e+00 | 1.685 | 1 | 6 | 92 |
| GO:0051607 | defense response to virus | 2.69e-01 | 1.00e+00 | 1.685 | 1 | 1 | 92 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 2.69e-01 | 1.00e+00 | 1.685 | 1 | 6 | 92 |
| GO:0006260 | DNA replication | 2.74e-01 | 1.00e+00 | 1.654 | 1 | 9 | 94 |
| GO:0034220 | ion transmembrane transport | 2.79e-01 | 1.00e+00 | 1.624 | 1 | 2 | 96 |
| GO:0007601 | visual perception | 2.81e-01 | 1.00e+00 | 1.609 | 1 | 2 | 97 |
| GO:0006457 | protein folding | 2.81e-01 | 1.00e+00 | 1.609 | 1 | 5 | 97 |
| GO:0016323 | basolateral plasma membrane | 3.03e-01 | 1.00e+00 | 1.481 | 1 | 3 | 106 |
| GO:0000790 | nuclear chromatin | 3.10e-01 | 1.00e+00 | 1.441 | 1 | 9 | 109 |
| GO:0006397 | mRNA processing | 3.12e-01 | 1.00e+00 | 1.428 | 1 | 12 | 110 |
| GO:0042981 | regulation of apoptotic process | 3.17e-01 | 1.00e+00 | 1.402 | 1 | 4 | 112 |
| GO:0043547 | positive regulation of GTPase activity | 3.17e-01 | 1.00e+00 | 1.402 | 1 | 2 | 112 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.21e-01 | 1.00e+00 | 0.795 | 2 | 23 | 341 |
| GO:0007049 | cell cycle | 3.24e-01 | 1.00e+00 | 1.364 | 1 | 8 | 115 |
| GO:0003924 | GTPase activity | 3.33e-01 | 1.00e+00 | 1.314 | 1 | 7 | 119 |
| GO:0042802 | identical protein binding | 3.37e-01 | 1.00e+00 | 0.741 | 2 | 15 | 354 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 3.47e-01 | 1.00e+00 | 1.243 | 1 | 4 | 125 |
| GO:0005524 | ATP binding | 3.56e-01 | 1.00e+00 | 0.408 | 4 | 37 | 892 |
| GO:0006184 | GTP catabolic process | 3.58e-01 | 1.00e+00 | 1.187 | 1 | 7 | 130 |
| GO:0003714 | transcription corepressor activity | 3.73e-01 | 1.00e+00 | 1.111 | 1 | 5 | 137 |
| GO:0044212 | transcription regulatory region DNA binding | 3.86e-01 | 1.00e+00 | 1.049 | 1 | 13 | 143 |
| GO:0046872 | metal ion binding | 3.89e-01 | 1.00e+00 | 0.342 | 4 | 24 | 934 |
| GO:0005622 | intracellular | 3.90e-01 | 1.00e+00 | 1.029 | 1 | 2 | 145 |
| GO:0004871 | signal transducer activity | 3.96e-01 | 1.00e+00 | 1.000 | 1 | 4 | 148 |
| GO:0005886 | plasma membrane | 3.98e-01 | 1.00e+00 | 0.215 | 7 | 46 | 1784 |
| GO:0016021 | integral component of membrane | 4.14e-01 | 1.00e+00 | 0.218 | 6 | 19 | 1526 |
| GO:0005874 | microtubule | 4.27e-01 | 1.00e+00 | 0.860 | 1 | 10 | 163 |
| GO:0016071 | mRNA metabolic process | 4.40e-01 | 1.00e+00 | 0.800 | 1 | 10 | 170 |
| GO:0019221 | cytokine-mediated signaling pathway | 4.44e-01 | 1.00e+00 | 0.783 | 1 | 10 | 172 |
| GO:0008270 | zinc ion binding | 4.50e-01 | 1.00e+00 | 0.288 | 3 | 39 | 727 |
| GO:0000166 | nucleotide binding | 4.52e-01 | 1.00e+00 | 0.750 | 1 | 13 | 176 |
| GO:0008152 | metabolic process | 4.63e-01 | 1.00e+00 | 0.701 | 1 | 6 | 182 |
| GO:0016567 | protein ubiquitination | 4.74e-01 | 1.00e+00 | 0.654 | 1 | 14 | 188 |
| GO:0016070 | RNA metabolic process | 4.74e-01 | 1.00e+00 | 0.654 | 1 | 10 | 188 |
| GO:0005525 | GTP binding | 4.90e-01 | 1.00e+00 | 0.587 | 1 | 12 | 197 |
| GO:0006955 | immune response | 4.95e-01 | 1.00e+00 | 0.565 | 1 | 2 | 200 |
| GO:0043065 | positive regulation of apoptotic process | 4.99e-01 | 1.00e+00 | 0.551 | 1 | 7 | 202 |
| GO:0006351 | transcription, DNA-templated | 5.01e-01 | 1.00e+00 | 0.138 | 4 | 47 | 1076 |
| GO:0006281 | DNA repair | 5.01e-01 | 1.00e+00 | 0.544 | 1 | 24 | 203 |
| GO:0015031 | protein transport | 5.01e-01 | 1.00e+00 | 0.544 | 1 | 10 | 203 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 5.01e-01 | 1.00e+00 | 0.544 | 1 | 14 | 203 |
| GO:0030154 | cell differentiation | 5.02e-01 | 1.00e+00 | 0.537 | 1 | 5 | 204 |
| GO:0008134 | transcription factor binding | 5.11e-01 | 1.00e+00 | 0.502 | 1 | 10 | 209 |
| GO:0019899 | enzyme binding | 5.13e-01 | 1.00e+00 | 0.495 | 1 | 7 | 210 |
| GO:0007275 | multicellular organismal development | 5.19e-01 | 1.00e+00 | 0.468 | 1 | 9 | 214 |
| GO:0006200 | ATP catabolic process | 5.28e-01 | 1.00e+00 | 0.434 | 1 | 13 | 219 |
| GO:0006508 | proteolysis | 5.96e-01 | 1.00e+00 | 0.165 | 1 | 11 | 264 |
| GO:0046982 | protein heterodimerization activity | 5.97e-01 | 1.00e+00 | 0.159 | 1 | 11 | 265 |
| GO:0005730 | nucleolus | 6.03e-01 | 1.00e+00 | -0.040 | 4 | 74 | 1217 |
| GO:0007165 | signal transduction | 6.27e-01 | 1.00e+00 | -0.062 | 2 | 24 | 618 |
| GO:0009986 | cell surface | 6.32e-01 | 1.00e+00 | 0.024 | 1 | 5 | 291 |
| GO:0000278 | mitotic cell cycle | 6.61e-01 | 1.00e+00 | -0.086 | 1 | 28 | 314 |
| GO:0055085 | transmembrane transport | 6.78e-01 | 1.00e+00 | -0.153 | 1 | 13 | 329 |
| GO:0007596 | blood coagulation | 6.89e-01 | 1.00e+00 | -0.196 | 1 | 13 | 339 |
| GO:0005634 | nucleus | 7.01e-01 | 1.00e+00 | -0.134 | 10 | 158 | 3246 |
| GO:0005509 | calcium ion binding | 7.02e-01 | 1.00e+00 | -0.246 | 1 | 9 | 351 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 7.28e-01 | 1.00e+00 | -0.349 | 1 | 21 | 377 |
| GO:0005783 | endoplasmic reticulum | 7.32e-01 | 1.00e+00 | -0.365 | 1 | 12 | 381 |
| GO:0006915 | apoptotic process | 7.59e-01 | 1.00e+00 | -0.474 | 1 | 15 | 411 |
| GO:0005794 | Golgi apparatus | 7.62e-01 | 1.00e+00 | -0.488 | 1 | 6 | 415 |
| GO:0045087 | innate immune response | 7.91e-01 | 1.00e+00 | -0.611 | 1 | 12 | 452 |
| GO:0003677 | DNA binding | 8.44e-01 | 1.00e+00 | -0.678 | 2 | 52 | 947 |
| GO:0005887 | integral component of plasma membrane | 8.65e-01 | 1.00e+00 | -0.958 | 1 | 8 | 575 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 8.69e-01 | 1.00e+00 | -0.976 | 1 | 22 | 582 |
| GO:0005615 | extracellular space | 8.98e-01 | 1.00e+00 | -1.138 | 1 | 15 | 651 |
| GO:0005576 | extracellular region | 9.02e-01 | 1.00e+00 | -1.164 | 1 | 15 | 663 |
| GO:0006355 | regulation of transcription, DNA-templated | 9.19e-01 | 1.00e+00 | -1.273 | 1 | 31 | 715 |
| GO:0070062 | extracellular vesicular exosome | 9.33e-01 | 1.00e+00 | -0.886 | 3 | 51 | 1641 |
| GO:0044281 | small molecule metabolic process | 9.50e-01 | 1.00e+00 | -1.512 | 1 | 35 | 844 |