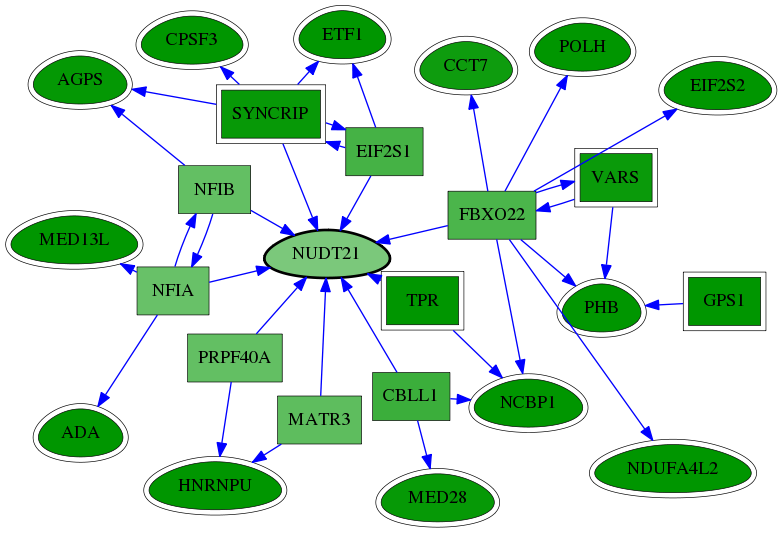
| GO:0003723 | RNA binding | 1.98e-06 | 1.94e-02 | 3.469 | 7 | 22 | 247 |
| GO:0008380 | RNA splicing | 2.88e-06 | 2.82e-02 | 3.828 | 6 | 21 | 165 |
| GO:0044822 | poly(A) RNA binding | 1.33e-05 | 1.30e-01 | 2.289 | 10 | 45 | 799 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.54e-05 | 1.50e-01 | 3.931 | 5 | 15 | 128 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 1.88e-05 | 1.84e-01 | 8.025 | 2 | 2 | 3 |
| GO:0010467 | gene expression | 3.63e-05 | 3.55e-01 | 2.546 | 8 | 49 | 535 |
| GO:0030529 | ribonucleoprotein complex | 5.11e-05 | 4.99e-01 | 4.252 | 4 | 5 | 82 |
| GO:0070934 | CRD-mediated mRNA stabilization | 6.26e-05 | 6.12e-01 | 7.288 | 2 | 2 | 5 |
| GO:0031124 | mRNA 3'-end processing | 9.19e-05 | 8.98e-01 | 5.065 | 3 | 4 | 35 |
| GO:0070937 | CRD-mediated mRNA stability complex | 9.38e-05 | 9.16e-01 | 7.025 | 2 | 2 | 6 |
| GO:0006369 | termination of RNA polymerase II transcription | 1.18e-04 | 1.00e+00 | 4.947 | 3 | 5 | 38 |
| GO:0006379 | mRNA cleavage | 2.24e-04 | 1.00e+00 | 6.440 | 2 | 2 | 9 |
| GO:0043022 | ribosome binding | 8.36e-04 | 1.00e+00 | 5.522 | 2 | 2 | 17 |
| GO:0001649 | osteoblast differentiation | 8.52e-04 | 1.00e+00 | 3.985 | 3 | 5 | 74 |
| GO:0006378 | mRNA polyadenylation | 9.39e-04 | 1.00e+00 | 5.440 | 2 | 3 | 18 |
| GO:0005654 | nucleoplasm | 1.10e-03 | 1.00e+00 | 1.835 | 8 | 68 | 876 |
| GO:0006366 | transcription from RNA polymerase II promoter | 1.50e-03 | 1.00e+00 | 2.518 | 5 | 23 | 341 |
| GO:0006260 | DNA replication | 1.70e-03 | 1.00e+00 | 3.640 | 3 | 9 | 94 |
| GO:0016592 | mediator complex | 1.82e-03 | 1.00e+00 | 4.966 | 2 | 6 | 25 |
| GO:0016020 | membrane | 2.10e-03 | 1.00e+00 | 1.542 | 9 | 56 | 1207 |
| GO:0005515 | protein binding | 2.50e-03 | 1.00e+00 | 0.770 | 18 | 198 | 4124 |
| GO:0004832 | valine-tRNA ligase activity | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:1901673 | regulation of spindle assembly involved in mitosis | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0043578 | nuclear matrix organization | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0010965 | regulation of mitotic sister chromatid separation | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0008609 | alkylglycerone-phosphate synthase activity | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0006404 | RNA import into nucleus | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0006157 | deoxyadenosine catabolic process | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0006438 | valyl-tRNA aminoacylation | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0046111 | xanthine biosynthetic process | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0032261 | purine nucleotide salvage | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0060169 | negative regulation of adenosine receptor signaling pathway | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0001883 | purine nucleoside binding | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0016149 | translation release factor activity, codon specific | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0021740 | principal sensory nucleus of trigeminal nerve development | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0008079 | translation termination factor activity | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0060407 | negative regulation of penile erection | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0031453 | positive regulation of heterochromatin assembly | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0061141 | lung ciliated cell differentiation | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0070256 | negative regulation of mucus secretion | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 1 |
| GO:0003743 | translation initiation factor activity | 3.97e-03 | 1.00e+00 | 4.400 | 2 | 8 | 37 |
| GO:0006396 | RNA processing | 4.62e-03 | 1.00e+00 | 4.288 | 2 | 4 | 40 |
| GO:0002314 | germinal center B cell differentiation | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0031990 | mRNA export from nucleus in response to heat stress | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0006154 | adenosine catabolic process | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0042306 | regulation of protein import into nucleus | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0005846 | nuclear cap binding complex | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0002176 | male germ cell proliferation | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0070840 | dynein complex binding | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0046103 | inosine biosynthetic process | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0008611 | ether lipid biosynthetic process | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0046832 | negative regulation of RNA export from nucleus | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0000189 | MAPK import into nucleus | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0048742 | regulation of skeletal muscle fiber development | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0043558 | regulation of translational initiation in response to stress | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0003747 | translation release factor activity | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 2 | 2 |
| GO:2000791 | negative regulation of mesenchymal cell proliferation involved in lung development | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0060205 | cytoplasmic membrane-bounded vesicle lumen | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 2 |
| GO:0006406 | mRNA export from nucleus | 5.57e-03 | 1.00e+00 | 4.150 | 2 | 7 | 44 |
| GO:0008762 | UDP-N-acetylmuramate dehydrogenase activity | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0043103 | hypoxanthine salvage | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0002686 | negative regulation of leukocyte migration | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0033632 | regulation of cell-cell adhesion mediated by integrin | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0046061 | dATP catabolic process | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0006405 | RNA export from nucleus | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0002636 | positive regulation of germinal center formation | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0010793 | regulation of mRNA export from nucleus | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 3 |
| GO:0003729 | mRNA binding | 9.21e-03 | 1.00e+00 | 3.777 | 2 | 4 | 57 |
| GO:0042826 | histone deacetylase binding | 9.53e-03 | 1.00e+00 | 3.752 | 2 | 5 | 58 |
| GO:0097452 | GAIT complex | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 2 | 4 |
| GO:0005849 | mRNA cleavage factor complex | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0021960 | anterior commissure morphogenesis | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0044300 | cerebellar mossy fiber | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0060486 | Clara cell differentiation | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 2 | 4 |
| GO:0051151 | negative regulation of smooth muscle cell differentiation | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0050862 | positive regulation of T cell receptor signaling pathway | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0045292 | mRNA cis splicing, via spliceosome | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0031442 | positive regulation of mRNA 3'-end processing | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0006290 | pyrimidine dimer repair | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 2 | 4 |
| GO:0006449 | regulation of translational termination | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0002906 | negative regulation of mature B cell apoptotic process | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0006412 | translation | 1.02e-02 | 1.00e+00 | 2.719 | 3 | 12 | 178 |
| GO:0006999 | nuclear pore organization | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0070849 | response to epidermal growth factor | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0044615 | nuclear pore nuclear basket | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0001821 | histamine secretion | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 4 |
| GO:0071013 | catalytic step 2 spliceosome | 1.05e-02 | 1.00e+00 | 3.679 | 2 | 5 | 61 |
| GO:0044267 | cellular protein metabolic process | 1.15e-02 | 1.00e+00 | 2.150 | 4 | 14 | 352 |
| GO:0008610 | lipid biosynthetic process | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0046825 | regulation of protein export from nucleus | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0004000 | adenosine deaminase activity | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0002161 | aminoacyl-tRNA editing activity | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0004521 | endoribonuclease activity | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0070244 | negative regulation of thymocyte apoptotic process | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 2 | 5 |
| GO:0006398 | histone mRNA 3'-end processing | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0060510 | Type II pneumocyte differentiation | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0043495 | protein anchor | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0006282 | regulation of DNA repair | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 2 | 5 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0042382 | paraspeckles | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 3 | 5 |
| GO:0033089 | positive regulation of T cell differentiation in thymus | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 5 |
| GO:0016363 | nuclear matrix | 1.48e-02 | 1.00e+00 | 3.420 | 2 | 10 | 73 |
| GO:0050847 | progesterone receptor signaling pathway | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 2 | 6 |
| GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 6 |
| GO:0060509 | Type I pneumocyte differentiation | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 6 |
| GO:0071679 | commissural neuron axon guidance | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 6 |
| GO:0045987 | positive regulation of smooth muscle contraction | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 6 |
| GO:0002199 | zona pellucida receptor complex | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 4 | 7 |
| GO:0008409 | 5'-3' exonuclease activity | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 2 | 7 |
| GO:0000339 | RNA cap binding | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 7 |
| GO:0033197 | response to vitamin E | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 7 |
| GO:0019079 | viral genome replication | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 7 |
| GO:0010460 | positive regulation of heart rate | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 7 |
| GO:0000731 | DNA synthesis involved in DNA repair | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 7 |
| GO:0006301 | postreplication repair | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 2 | 7 |
| GO:0048541 | Peyer's patch development | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 2 | 7 |
| GO:0005832 | chaperonin-containing T-complex | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 4 | 7 |
| GO:0005868 | cytoplasmic dynein complex | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 7 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1.93e-02 | 1.00e+00 | 3.217 | 2 | 5 | 84 |
| GO:0005845 | mRNA cap binding complex | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0042405 | nuclear inclusion body | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0051292 | nuclear pore complex assembly | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 4 | 8 |
| GO:0006450 | regulation of translational fidelity | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0071949 | FAD binding | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0010388 | cullin deneddylation | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 4 | 8 |
| GO:0035457 | cellular response to interferon-alpha | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0046638 | positive regulation of alpha-beta T cell differentiation | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0043101 | purine-containing compound salvage | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 1 | 8 |
| GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 2 | 9 |
| GO:0045947 | negative regulation of translational initiation | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 1 | 9 |
| GO:0071354 | cellular response to interleukin-6 | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 1 | 9 |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 3 | 9 |
| GO:0048566 | embryonic digestive tract development | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 1 | 9 |
| GO:0010225 | response to UV-C | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 2 | 9 |
| GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 2 | 9 |
| GO:0005739 | mitochondrion | 2.37e-02 | 1.00e+00 | 1.567 | 5 | 32 | 659 |
| GO:0005095 | GTPase inhibitor activity | 2.53e-02 | 1.00e+00 | 5.288 | 1 | 1 | 10 |
| GO:0008143 | poly(A) binding | 2.53e-02 | 1.00e+00 | 5.288 | 1 | 2 | 10 |
| GO:0032465 | regulation of cytokinesis | 2.53e-02 | 1.00e+00 | 5.288 | 1 | 1 | 10 |
| GO:0006413 | translational initiation | 2.62e-02 | 1.00e+00 | 2.980 | 2 | 6 | 99 |
| GO:0008334 | histone mRNA metabolic process | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 1 | 11 |
| GO:0010001 | glial cell differentiation | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 1 | 11 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 4 | 11 |
| GO:0006479 | protein methylation | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 2 | 11 |
| GO:0051019 | mitogen-activated protein kinase binding | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 1 | 11 |
| GO:0090316 | positive regulation of intracellular protein transport | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 1 | 11 |
| GO:0045807 | positive regulation of endocytosis | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 1 | 11 |
| GO:0005487 | nucleocytoplasmic transporter activity | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 2 | 11 |
| GO:0046827 | positive regulation of protein export from nucleus | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 1 | 11 |
| GO:0042307 | positive regulation of protein import into nucleus | 3.03e-02 | 1.00e+00 | 5.025 | 1 | 3 | 12 |
| GO:0034399 | nuclear periphery | 3.03e-02 | 1.00e+00 | 5.025 | 1 | 3 | 12 |
| GO:0006397 | mRNA processing | 3.19e-02 | 1.00e+00 | 2.828 | 2 | 12 | 110 |
| GO:0017091 | AU-rich element binding | 3.28e-02 | 1.00e+00 | 4.909 | 1 | 2 | 13 |
| GO:0007049 | cell cycle | 3.46e-02 | 1.00e+00 | 2.764 | 2 | 8 | 115 |
| GO:0030902 | hindbrain development | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 2 | 14 |
| GO:0070064 | proline-rich region binding | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 1 | 14 |
| GO:0016575 | histone deacetylation | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 2 | 14 |
| GO:0032839 | dendrite cytoplasm | 3.77e-02 | 1.00e+00 | 4.703 | 1 | 1 | 15 |
| GO:0008135 | translation factor activity, nucleic acid binding | 3.77e-02 | 1.00e+00 | 4.703 | 1 | 4 | 15 |
| GO:0000188 | inactivation of MAPK activity | 3.77e-02 | 1.00e+00 | 4.703 | 1 | 1 | 15 |
| GO:0007339 | binding of sperm to zona pellucida | 3.77e-02 | 1.00e+00 | 4.703 | 1 | 4 | 15 |
| GO:0030864 | cortical actin cytoskeleton | 4.02e-02 | 1.00e+00 | 4.610 | 1 | 1 | 16 |
| GO:0003887 | DNA-directed DNA polymerase activity | 4.02e-02 | 1.00e+00 | 4.610 | 1 | 2 | 16 |
| GO:0005782 | peroxisomal matrix | 4.02e-02 | 1.00e+00 | 4.610 | 1 | 1 | 16 |
| GO:0005634 | nucleus | 4.04e-02 | 1.00e+00 | 0.646 | 13 | 158 | 3246 |
| GO:0001829 | trophectodermal cell differentiation | 4.27e-02 | 1.00e+00 | 4.522 | 1 | 2 | 17 |
| GO:0050850 | positive regulation of calcium-mediated signaling | 4.27e-02 | 1.00e+00 | 4.522 | 1 | 1 | 17 |
| GO:0043278 | response to morphine | 4.27e-02 | 1.00e+00 | 4.522 | 1 | 2 | 17 |
| GO:0001077 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 4.44e-02 | 1.00e+00 | 2.565 | 2 | 4 | 132 |
| GO:0006611 | protein export from nucleus | 4.51e-02 | 1.00e+00 | 4.440 | 1 | 1 | 18 |
| GO:0031047 | gene silencing by RNA | 4.51e-02 | 1.00e+00 | 4.440 | 1 | 2 | 18 |
| GO:0001106 | RNA polymerase II transcription corepressor activity | 4.51e-02 | 1.00e+00 | 4.440 | 1 | 2 | 18 |
| GO:0005844 | polysome | 4.76e-02 | 1.00e+00 | 4.362 | 1 | 2 | 19 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 4.76e-02 | 1.00e+00 | 4.362 | 1 | 1 | 19 |
| GO:0015631 | tubulin binding | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 1 | 20 |
| GO:0006913 | nucleocytoplasmic transport | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 2 | 20 |
| GO:0071346 | cellular response to interferon-gamma | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 2 | 20 |
| GO:0034660 | ncRNA metabolic process | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 1 | 20 |
| GO:0005637 | nuclear inner membrane | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 1 | 20 |
| GO:0072686 | mitotic spindle | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 2 | 20 |
| GO:0071897 | DNA biosynthetic process | 5.00e-02 | 1.00e+00 | 4.288 | 1 | 2 | 20 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 5.07e-02 | 1.00e+00 | 2.460 | 2 | 7 | 142 |
| GO:0031647 | regulation of protein stability | 5.25e-02 | 1.00e+00 | 4.217 | 1 | 2 | 21 |
| GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 5.25e-02 | 1.00e+00 | 4.217 | 1 | 2 | 21 |
| GO:0006606 | protein import into nucleus | 5.49e-02 | 1.00e+00 | 4.150 | 1 | 3 | 22 |
| GO:0051084 | 'de novo' posttranslational protein folding | 5.49e-02 | 1.00e+00 | 4.150 | 1 | 4 | 22 |
| GO:0000387 | spliceosomal snRNP assembly | 5.49e-02 | 1.00e+00 | 4.150 | 1 | 1 | 22 |
| GO:0002062 | chondrocyte differentiation | 5.49e-02 | 1.00e+00 | 4.150 | 1 | 2 | 22 |
| GO:0034605 | cellular response to heat | 5.49e-02 | 1.00e+00 | 4.150 | 1 | 1 | 22 |
| GO:0051262 | protein tetramerization | 5.73e-02 | 1.00e+00 | 4.086 | 1 | 2 | 23 |
| GO:0043392 | negative regulation of DNA binding | 5.73e-02 | 1.00e+00 | 4.086 | 1 | 1 | 23 |
| GO:0006370 | 7-methylguanosine mRNA capping | 5.97e-02 | 1.00e+00 | 4.025 | 1 | 5 | 24 |
| GO:0006144 | purine nucleobase metabolic process | 5.97e-02 | 1.00e+00 | 4.025 | 1 | 2 | 24 |
| GO:0008180 | COP9 signalosome | 5.97e-02 | 1.00e+00 | 4.025 | 1 | 6 | 24 |
| GO:0007162 | negative regulation of cell adhesion | 5.97e-02 | 1.00e+00 | 4.025 | 1 | 2 | 24 |
| GO:0001104 | RNA polymerase II transcription cofactor activity | 5.97e-02 | 1.00e+00 | 4.025 | 1 | 4 | 24 |
| GO:0010827 | regulation of glucose transport | 6.21e-02 | 1.00e+00 | 3.966 | 1 | 7 | 25 |
| GO:0007094 | mitotic spindle assembly checkpoint | 6.21e-02 | 1.00e+00 | 3.966 | 1 | 4 | 25 |
| GO:0009267 | cellular response to starvation | 6.46e-02 | 1.00e+00 | 3.909 | 1 | 1 | 26 |
| GO:0001890 | placenta development | 6.46e-02 | 1.00e+00 | 3.909 | 1 | 2 | 26 |
| GO:0010494 | cytoplasmic stress granule | 6.46e-02 | 1.00e+00 | 3.909 | 1 | 4 | 26 |
| GO:0048286 | lung alveolus development | 6.70e-02 | 1.00e+00 | 3.855 | 1 | 1 | 27 |
| GO:0031072 | heat shock protein binding | 6.94e-02 | 1.00e+00 | 3.802 | 1 | 1 | 28 |
| GO:0016071 | mRNA metabolic process | 6.96e-02 | 1.00e+00 | 2.200 | 2 | 10 | 170 |
| GO:0006418 | tRNA aminoacylation for protein translation | 7.17e-02 | 1.00e+00 | 3.752 | 1 | 4 | 29 |
| GO:0005778 | peroxisomal membrane | 7.17e-02 | 1.00e+00 | 3.752 | 1 | 3 | 29 |
| GO:0006446 | regulation of translational initiation | 7.17e-02 | 1.00e+00 | 3.752 | 1 | 2 | 29 |
| GO:0000166 | nucleotide binding | 7.39e-02 | 1.00e+00 | 2.150 | 2 | 13 | 176 |
| GO:0019898 | extrinsic component of membrane | 7.41e-02 | 1.00e+00 | 3.703 | 1 | 1 | 30 |
| GO:0004842 | ubiquitin-protein transferase activity | 7.61e-02 | 1.00e+00 | 2.126 | 2 | 17 | 179 |
| GO:0007077 | mitotic nuclear envelope disassembly | 7.89e-02 | 1.00e+00 | 3.610 | 1 | 9 | 32 |
| GO:0042110 | T cell activation | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 2 | 33 |
| GO:0030890 | positive regulation of B cell proliferation | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 2 | 33 |
| GO:0017148 | negative regulation of translation | 8.12e-02 | 1.00e+00 | 3.565 | 1 | 5 | 33 |
| GO:0016070 | RNA metabolic process | 8.29e-02 | 1.00e+00 | 2.055 | 2 | 10 | 188 |
| GO:0014070 | response to organic cyclic compound | 8.36e-02 | 1.00e+00 | 3.522 | 1 | 2 | 34 |
| GO:0007254 | JNK cascade | 8.36e-02 | 1.00e+00 | 3.522 | 1 | 1 | 34 |
| GO:0044297 | cell body | 8.60e-02 | 1.00e+00 | 3.480 | 1 | 5 | 35 |
| GO:0050434 | positive regulation of viral transcription | 8.60e-02 | 1.00e+00 | 3.480 | 1 | 5 | 35 |
| GO:0008645 | hexose transport | 8.83e-02 | 1.00e+00 | 3.440 | 1 | 8 | 36 |
| GO:0042542 | response to hydrogen peroxide | 8.83e-02 | 1.00e+00 | 3.440 | 1 | 1 | 36 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 9.30e-02 | 1.00e+00 | 3.362 | 1 | 4 | 38 |
| GO:0016032 | viral process | 9.43e-02 | 1.00e+00 | 1.453 | 3 | 30 | 428 |
| GO:0019827 | stem cell maintenance | 9.53e-02 | 1.00e+00 | 3.324 | 1 | 4 | 39 |
| GO:0016787 | hydrolase activity | 9.76e-02 | 1.00e+00 | 3.288 | 1 | 3 | 40 |
| GO:0003684 | damaged DNA binding | 1.00e-01 | 1.00e+00 | 3.252 | 1 | 1 | 41 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 1.09e-01 | 1.00e+00 | 1.362 | 3 | 29 | 456 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 1.14e-01 | 1.00e+00 | 3.055 | 1 | 1 | 47 |
| GO:0000151 | ubiquitin ligase complex | 1.16e-01 | 1.00e+00 | 3.025 | 1 | 4 | 48 |
| GO:0015758 | glucose transport | 1.16e-01 | 1.00e+00 | 3.025 | 1 | 8 | 48 |
| GO:0050728 | negative regulation of inflammatory response | 1.18e-01 | 1.00e+00 | 2.995 | 1 | 1 | 49 |
| GO:0051301 | cell division | 1.18e-01 | 1.00e+00 | 2.995 | 1 | 2 | 49 |
| GO:0005777 | peroxisome | 1.21e-01 | 1.00e+00 | 2.966 | 1 | 2 | 50 |
| GO:0005643 | nuclear pore | 1.23e-01 | 1.00e+00 | 2.937 | 1 | 12 | 51 |
| GO:0055086 | nucleobase-containing small molecule metabolic process | 1.23e-01 | 1.00e+00 | 2.937 | 1 | 2 | 51 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 1.25e-01 | 1.00e+00 | 2.909 | 1 | 8 | 52 |
| GO:0000776 | kinetochore | 1.32e-01 | 1.00e+00 | 2.828 | 1 | 8 | 55 |
| GO:0008584 | male gonad development | 1.36e-01 | 1.00e+00 | 2.777 | 1 | 1 | 57 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 1.41e-01 | 1.00e+00 | 2.727 | 1 | 1 | 59 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 1.45e-01 | 1.00e+00 | 2.679 | 1 | 1 | 61 |
| GO:0006464 | cellular protein modification process | 1.45e-01 | 1.00e+00 | 2.679 | 1 | 4 | 61 |
| GO:0007010 | cytoskeleton organization | 1.45e-01 | 1.00e+00 | 2.679 | 1 | 4 | 61 |
| GO:0016337 | single organismal cell-cell adhesion | 1.47e-01 | 1.00e+00 | 2.655 | 1 | 3 | 62 |
| GO:0006415 | translational termination | 1.50e-01 | 1.00e+00 | 2.632 | 1 | 2 | 63 |
| GO:0030018 | Z disc | 1.52e-01 | 1.00e+00 | 2.610 | 1 | 3 | 64 |
| GO:0005829 | cytosol | 1.58e-01 | 1.00e+00 | 0.614 | 7 | 88 | 1787 |
| GO:0008360 | regulation of cell shape | 1.65e-01 | 1.00e+00 | 2.480 | 1 | 3 | 70 |
| GO:0051082 | unfolded protein binding | 1.65e-01 | 1.00e+00 | 2.480 | 1 | 5 | 70 |
| GO:0030308 | negative regulation of cell growth | 1.65e-01 | 1.00e+00 | 2.480 | 1 | 3 | 70 |
| GO:0001889 | liver development | 1.67e-01 | 1.00e+00 | 2.460 | 1 | 4 | 71 |
| GO:0009986 | cell surface | 1.70e-01 | 1.00e+00 | 1.425 | 2 | 5 | 291 |
| GO:0005215 | transporter activity | 1.90e-01 | 1.00e+00 | 2.252 | 1 | 5 | 82 |
| GO:0005635 | nuclear envelope | 1.92e-01 | 1.00e+00 | 2.235 | 1 | 6 | 83 |
| GO:0003690 | double-stranded DNA binding | 1.92e-01 | 1.00e+00 | 2.235 | 1 | 6 | 83 |
| GO:0005730 | nucleolus | 1.93e-01 | 1.00e+00 | 0.682 | 5 | 74 | 1217 |
| GO:0044255 | cellular lipid metabolic process | 2.05e-01 | 1.00e+00 | 2.134 | 1 | 2 | 89 |
| GO:0000209 | protein polyubiquitination | 2.11e-01 | 1.00e+00 | 2.086 | 1 | 6 | 92 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 2.11e-01 | 1.00e+00 | 2.086 | 1 | 6 | 92 |
| GO:0016477 | cell migration | 2.19e-01 | 1.00e+00 | 2.025 | 1 | 3 | 96 |
| GO:0006457 | protein folding | 2.21e-01 | 1.00e+00 | 2.010 | 1 | 5 | 97 |
| GO:0007568 | aging | 2.21e-01 | 1.00e+00 | 2.010 | 1 | 3 | 97 |
| GO:0005764 | lysosome | 2.27e-01 | 1.00e+00 | 1.966 | 1 | 2 | 100 |
| GO:0005198 | structural molecule activity | 2.29e-01 | 1.00e+00 | 1.951 | 1 | 4 | 101 |
| GO:0030335 | positive regulation of cell migration | 2.29e-01 | 1.00e+00 | 1.951 | 1 | 2 | 101 |
| GO:0042802 | identical protein binding | 2.29e-01 | 1.00e+00 | 1.142 | 2 | 15 | 354 |
| GO:0046777 | protein autophosphorylation | 2.39e-01 | 1.00e+00 | 1.882 | 1 | 3 | 106 |
| GO:0042981 | regulation of apoptotic process | 2.51e-01 | 1.00e+00 | 1.802 | 1 | 4 | 112 |
| GO:0001666 | response to hypoxia | 2.59e-01 | 1.00e+00 | 1.752 | 1 | 2 | 116 |
| GO:0016607 | nuclear speck | 2.70e-01 | 1.00e+00 | 1.679 | 1 | 8 | 122 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.76e-01 | 1.00e+00 | 0.713 | 3 | 31 | 715 |
| GO:0008270 | zinc ion binding | 2.84e-01 | 1.00e+00 | 0.689 | 3 | 39 | 727 |
| GO:0031965 | nuclear membrane | 2.85e-01 | 1.00e+00 | 1.587 | 1 | 8 | 130 |
| GO:0009897 | external side of plasma membrane | 2.87e-01 | 1.00e+00 | 1.576 | 1 | 3 | 131 |
| GO:0042803 | protein homodimerization activity | 3.00e-01 | 1.00e+00 | 0.868 | 2 | 13 | 428 |
| GO:0044212 | transcription regulatory region DNA binding | 3.09e-01 | 1.00e+00 | 1.450 | 1 | 13 | 143 |
| GO:0016874 | ligase activity | 3.14e-01 | 1.00e+00 | 1.420 | 1 | 11 | 146 |
| GO:0001701 | in utero embryonic development | 3.28e-01 | 1.00e+00 | 1.343 | 1 | 7 | 154 |
| GO:0005874 | microtubule | 3.44e-01 | 1.00e+00 | 1.261 | 1 | 10 | 163 |
| GO:0007067 | mitotic nuclear division | 3.47e-01 | 1.00e+00 | 1.243 | 1 | 9 | 165 |
| GO:0005737 | cytoplasm | 3.55e-01 | 1.00e+00 | 0.247 | 8 | 127 | 2633 |
| GO:0043025 | neuronal cell body | 3.57e-01 | 1.00e+00 | 1.192 | 1 | 3 | 171 |
| GO:0019221 | cytokine-mediated signaling pathway | 3.59e-01 | 1.00e+00 | 1.183 | 1 | 10 | 172 |
| GO:0044281 | small molecule metabolic process | 3.69e-01 | 1.00e+00 | 0.473 | 3 | 35 | 844 |
| GO:0003779 | actin binding | 3.71e-01 | 1.00e+00 | 1.126 | 1 | 3 | 179 |
| GO:0005975 | carbohydrate metabolic process | 3.82e-01 | 1.00e+00 | 1.070 | 1 | 10 | 186 |
| GO:0016567 | protein ubiquitination | 3.85e-01 | 1.00e+00 | 1.055 | 1 | 14 | 188 |
| GO:0005743 | mitochondrial inner membrane | 4.00e-01 | 1.00e+00 | 0.987 | 1 | 9 | 197 |
| GO:0005524 | ATP binding | 4.03e-01 | 1.00e+00 | 0.394 | 3 | 37 | 892 |
| GO:0006281 | DNA repair | 4.09e-01 | 1.00e+00 | 0.944 | 1 | 24 | 203 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 4.09e-01 | 1.00e+00 | 0.944 | 1 | 14 | 203 |
| GO:0008134 | transcription factor binding | 4.18e-01 | 1.00e+00 | 0.902 | 1 | 10 | 209 |
| GO:0019899 | enzyme binding | 4.20e-01 | 1.00e+00 | 0.895 | 1 | 7 | 210 |
| GO:0005813 | centrosome | 4.30e-01 | 1.00e+00 | 0.848 | 1 | 10 | 217 |
| GO:0030054 | cell junction | 4.32e-01 | 1.00e+00 | 0.841 | 1 | 5 | 218 |
| GO:0043231 | intracellular membrane-bounded organelle | 4.32e-01 | 1.00e+00 | 0.841 | 1 | 8 | 218 |
| GO:0046872 | metal ion binding | 4.33e-01 | 1.00e+00 | 0.327 | 3 | 24 | 934 |
| GO:0042493 | response to drug | 4.38e-01 | 1.00e+00 | 0.815 | 1 | 4 | 222 |
| GO:0007155 | cell adhesion | 4.69e-01 | 1.00e+00 | 0.679 | 1 | 8 | 244 |
| GO:0003682 | chromatin binding | 4.80e-01 | 1.00e+00 | 0.632 | 1 | 15 | 252 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 4.98e-01 | 1.00e+00 | 0.279 | 2 | 29 | 644 |
| GO:0008285 | negative regulation of cell proliferation | 5.13e-01 | 1.00e+00 | 0.496 | 1 | 11 | 277 |
| GO:0055114 | oxidation-reduction process | 5.52e-01 | 1.00e+00 | 0.343 | 1 | 9 | 308 |
| GO:0000278 | mitotic cell cycle | 5.59e-01 | 1.00e+00 | 0.315 | 1 | 28 | 314 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 5.62e-01 | 1.00e+00 | 0.301 | 1 | 17 | 317 |
| GO:0055085 | transmembrane transport | 5.76e-01 | 1.00e+00 | 0.248 | 1 | 13 | 329 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 6.27e-01 | 1.00e+00 | 0.051 | 1 | 21 | 377 |
| GO:0005783 | endoplasmic reticulum | 6.31e-01 | 1.00e+00 | 0.036 | 1 | 12 | 381 |
| GO:0003677 | DNA binding | 7.13e-01 | 1.00e+00 | -0.278 | 2 | 52 | 947 |
| GO:0006351 | transcription, DNA-templated | 7.79e-01 | 1.00e+00 | -0.462 | 2 | 47 | 1076 |
| GO:0005887 | integral component of plasma membrane | 7.81e-01 | 1.00e+00 | -0.558 | 1 | 8 | 575 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 7.85e-01 | 1.00e+00 | -0.575 | 1 | 22 | 582 |
| GO:0007165 | signal transduction | 8.05e-01 | 1.00e+00 | -0.662 | 1 | 24 | 618 |
| GO:0070062 | extracellular vesicular exosome | 8.16e-01 | 1.00e+00 | -0.486 | 3 | 51 | 1641 |
| GO:0005615 | extracellular space | 8.22e-01 | 1.00e+00 | -0.737 | 1 | 15 | 651 |
| GO:0005886 | plasma membrane | 9.58e-01 | 1.00e+00 | -1.191 | 2 | 46 | 1784 |