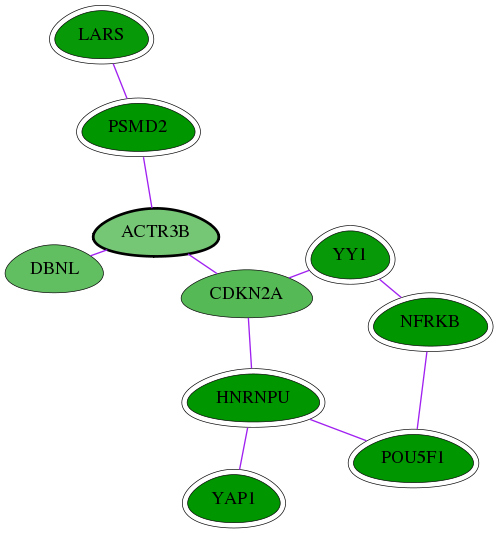
| GO:0031011 | Ino80 complex | 3.92e-05 | 5.65e-01 | 7.687 | 2 | 6 | 14 |
| GO:0044212 | transcription regulatory region DNA binding | 1.75e-04 | 1.00e+00 | 4.687 | 3 | 17 | 168 |
| GO:0005654 | nucleoplasm | 4.31e-04 | 1.00e+00 | 2.737 | 5 | 64 | 1082 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0003677 | DNA binding | 7.48e-04 | 1.00e+00 | 2.566 | 5 | 49 | 1218 |
| GO:0010467 | gene expression | 7.70e-04 | 1.00e+00 | 3.108 | 4 | 45 | 669 |
| GO:0034696 | response to prostaglandin F | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0006429 | leucyl-tRNA aminoacylation | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0060242 | contact inhibition | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0006403 | RNA localization | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0004823 | leucine-tRNA ligase activity | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0055105 | ubiquitin-protein transferase inhibitor activity | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0035985 | senescence-associated heterochromatin focus | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0035986 | senescence-associated heterochromatin focus assembly | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:2000111 | positive regulation of macrophage apoptotic process | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0005515 | protein binding | 2.41e-03 | 1.00e+00 | 1.108 | 9 | 198 | 6024 |
| GO:0071800 | podosome assembly | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:2000774 | positive regulation of cellular senescence | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0000209 | protein polyubiquitination | 2.77e-03 | 1.00e+00 | 4.636 | 2 | 3 | 116 |
| GO:0033088 | negative regulation of immature T cell proliferation in thymus | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0005829 | cytosol | 2.97e-03 | 1.00e+00 | 1.794 | 6 | 86 | 2496 |
| GO:0001714 | endodermal cell fate specification | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 2 | 5 |
| GO:1902510 | regulation of apoptotic DNA fragmentation | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 2 | 5 |
| GO:0097371 | MDM2/MDM4 family protein binding | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0070937 | CRD-mediated mRNA stability complex | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 3 | 6 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 4.52e-03 | 1.00e+00 | 4.275 | 2 | 3 | 149 |
| GO:0006974 | cellular response to DNA damage stimulus | 4.64e-03 | 1.00e+00 | 4.256 | 2 | 7 | 151 |
| GO:0046825 | regulation of protein export from nucleus | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0002161 | aminoacyl-tRNA editing activity | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0005667 | transcription factor complex | 6.18e-03 | 1.00e+00 | 4.043 | 2 | 17 | 175 |
| GO:0010225 | response to UV-C | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 2 | 9 |
| GO:0006450 | regulation of translational fidelity | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 2 | 9 |
| GO:0048103 | somatic stem cell division | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0000400 | four-way junction DNA binding | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0003714 | transcription corepressor activity | 6.39e-03 | 1.00e+00 | 4.018 | 2 | 11 | 178 |
| GO:0035413 | positive regulation of catenin import into nucleus | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 2 | 10 |
| GO:0090399 | replicative senescence | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 1 | 10 |
| GO:0033235 | positive regulation of protein sumoylation | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 1 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 4 | 11 |
| GO:0001953 | negative regulation of cell-matrix adhesion | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 12 |
| GO:0005838 | proteasome regulatory particle | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 12 |
| GO:0035198 | miRNA binding | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0071480 | cellular response to gamma radiation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 13 |
| GO:0030889 | negative regulation of B cell proliferation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0030234 | enzyme regulator activity | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 13 |
| GO:0030665 | clathrin-coated vesicle membrane | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 3 | 13 |
| GO:0001824 | blastocyst development | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 15 |
| GO:0042176 | regulation of protein catabolic process | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 2 | 16 |
| GO:0051276 | chromosome organization | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 2 | 16 |
| GO:0048593 | camera-type eye morphogenesis | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 16 |
| GO:0002102 | podosome | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 16 |
| GO:0003713 | transcription coactivator activity | 1.11e-02 | 1.00e+00 | 3.605 | 2 | 24 | 237 |
| GO:0008134 | transcription factor binding | 1.15e-02 | 1.00e+00 | 3.575 | 2 | 18 | 242 |
| GO:0016601 | Rac protein signal transduction | 1.17e-02 | 1.00e+00 | 6.407 | 1 | 1 | 17 |
| GO:0022624 | proteasome accessory complex | 1.17e-02 | 1.00e+00 | 6.407 | 1 | 1 | 17 |
| GO:0090398 | cellular senescence | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 18 |
| GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1.31e-02 | 1.00e+00 | 6.246 | 1 | 2 | 19 |
| GO:0071158 | positive regulation of cell cycle arrest | 1.31e-02 | 1.00e+00 | 6.246 | 1 | 2 | 19 |
| GO:0003779 | actin binding | 1.38e-02 | 1.00e+00 | 3.439 | 2 | 10 | 266 |
| GO:0005634 | nucleus | 1.44e-02 | 1.00e+00 | 1.147 | 7 | 159 | 4559 |
| GO:0042326 | negative regulation of phosphorylation | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 21 |
| GO:0008637 | apoptotic mitochondrial changes | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 2 | 21 |
| GO:0031648 | protein destabilization | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 21 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 2 | 22 |
| GO:0051059 | NF-kappaB binding | 1.72e-02 | 1.00e+00 | 5.850 | 1 | 3 | 25 |
| GO:0031519 | PcG protein complex | 1.72e-02 | 1.00e+00 | 5.850 | 1 | 1 | 25 |
| GO:0035329 | hippo signaling | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 5 | 26 |
| GO:0034644 | cellular response to UV | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 1 | 30 |
| GO:0031647 | regulation of protein stability | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 4 | 30 |
| GO:0048812 | neuron projection morphogenesis | 2.13e-02 | 1.00e+00 | 5.540 | 1 | 1 | 31 |
| GO:0070534 | protein K63-linked ubiquitination | 2.13e-02 | 1.00e+00 | 5.540 | 1 | 1 | 31 |
| GO:0016604 | nuclear body | 2.13e-02 | 1.00e+00 | 5.540 | 1 | 3 | 31 |
| GO:0002376 | immune system process | 2.20e-02 | 1.00e+00 | 5.494 | 1 | 3 | 32 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 2.20e-02 | 1.00e+00 | 5.494 | 1 | 2 | 32 |
| GO:0003723 | RNA binding | 2.22e-02 | 1.00e+00 | 3.076 | 2 | 18 | 342 |
| GO:0007416 | synapse assembly | 2.27e-02 | 1.00e+00 | 5.450 | 1 | 3 | 33 |
| GO:0005737 | cytoplasm | 2.44e-02 | 1.00e+00 | 1.200 | 6 | 124 | 3767 |
| GO:0007257 | activation of JUN kinase activity | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 5 | 36 |
| GO:0008047 | enzyme activator activity | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 3 | 39 |
| GO:0035019 | somatic stem cell maintenance | 2.74e-02 | 1.00e+00 | 5.172 | 1 | 6 | 40 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.74e-02 | 1.00e+00 | 2.118 | 3 | 43 | 997 |
| GO:0000278 | mitotic cell cycle | 2.86e-02 | 1.00e+00 | 2.883 | 2 | 15 | 391 |
| GO:0006418 | tRNA aminoacylation for protein translation | 2.88e-02 | 1.00e+00 | 5.102 | 1 | 3 | 42 |
| GO:0044822 | poly(A) RNA binding | 3.18e-02 | 1.00e+00 | 2.035 | 3 | 50 | 1056 |
| GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 3.21e-02 | 1.00e+00 | 4.940 | 1 | 2 | 47 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.25e-02 | 1.00e+00 | 2.783 | 2 | 30 | 419 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 1 | 50 |
| GO:0009611 | response to wounding | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 5 | 52 |
| GO:0000724 | double-strand break repair via homologous recombination | 3.75e-02 | 1.00e+00 | 4.713 | 1 | 2 | 55 |
| GO:0002039 | p53 binding | 3.75e-02 | 1.00e+00 | 4.713 | 1 | 4 | 55 |
| GO:0000502 | proteasome complex | 3.95e-02 | 1.00e+00 | 4.636 | 1 | 3 | 58 |
| GO:0006396 | RNA processing | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 5 | 59 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 5 | 59 |
| GO:0042995 | cell projection | 4.08e-02 | 1.00e+00 | 4.587 | 1 | 3 | 60 |
| GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 3 | 63 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 4.35e-02 | 1.00e+00 | 4.494 | 1 | 1 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 2 | 65 |
| GO:0006310 | DNA recombination | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 7 | 65 |
| GO:0006469 | negative regulation of protein kinase activity | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 4 | 65 |
| GO:0010468 | regulation of gene expression | 4.62e-02 | 1.00e+00 | 4.407 | 1 | 2 | 68 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.75e-02 | 1.00e+00 | 4.365 | 1 | 2 | 70 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 4.88e-02 | 1.00e+00 | 4.324 | 1 | 1 | 72 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 4 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 2 | 74 |
| GO:0002020 | protease binding | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 6 | 74 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 1 | 75 |
| GO:0007265 | Ras protein signal transduction | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 3 | 75 |
| GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 5.21e-02 | 1.00e+00 | 4.227 | 1 | 3 | 77 |
| GO:0071013 | catalytic step 2 spliceosome | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 7 | 78 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 5.35e-02 | 1.00e+00 | 4.190 | 1 | 2 | 79 |
| GO:0005524 | ATP binding | 5.40e-02 | 1.00e+00 | 1.737 | 3 | 31 | 1298 |
| GO:0001726 | ruffle | 5.41e-02 | 1.00e+00 | 4.172 | 1 | 4 | 80 |
| GO:0006915 | apoptotic process | 5.42e-02 | 1.00e+00 | 2.378 | 2 | 12 | 555 |
| GO:0009653 | anatomical structure morphogenesis | 5.61e-02 | 1.00e+00 | 4.119 | 1 | 2 | 83 |
| GO:0009952 | anterior/posterior pattern specification | 5.61e-02 | 1.00e+00 | 4.119 | 1 | 4 | 83 |
| GO:0050821 | protein stabilization | 5.94e-02 | 1.00e+00 | 4.035 | 1 | 3 | 88 |
| GO:0016363 | nuclear matrix | 6.14e-02 | 1.00e+00 | 3.986 | 1 | 6 | 91 |
| GO:0051015 | actin filament binding | 6.20e-02 | 1.00e+00 | 3.971 | 1 | 3 | 92 |
| GO:0001649 | osteoblast differentiation | 6.20e-02 | 1.00e+00 | 3.971 | 1 | 5 | 92 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 6.20e-02 | 1.00e+00 | 3.971 | 1 | 1 | 92 |
| GO:0006364 | rRNA processing | 6.33e-02 | 1.00e+00 | 3.940 | 1 | 3 | 94 |
| GO:0005938 | cell cortex | 6.98e-02 | 1.00e+00 | 3.794 | 1 | 5 | 104 |
| GO:0014069 | postsynaptic density | 7.05e-02 | 1.00e+00 | 3.780 | 1 | 1 | 105 |
| GO:0006897 | endocytosis | 7.11e-02 | 1.00e+00 | 3.766 | 1 | 7 | 106 |
| GO:0030308 | negative regulation of cell growth | 7.37e-02 | 1.00e+00 | 3.713 | 1 | 6 | 110 |
| GO:0030529 | ribonucleoprotein complex | 7.50e-02 | 1.00e+00 | 3.687 | 1 | 5 | 112 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 8.15e-02 | 1.00e+00 | 2.045 | 2 | 39 | 699 |
| GO:0007050 | cell cycle arrest | 8.27e-02 | 1.00e+00 | 3.540 | 1 | 5 | 124 |
| GO:0030027 | lamellipodium | 8.34e-02 | 1.00e+00 | 3.528 | 1 | 7 | 125 |
| GO:0042981 | regulation of apoptotic process | 9.93e-02 | 1.00e+00 | 3.265 | 1 | 4 | 150 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 1.00e-01 | 1.00e+00 | 1.870 | 2 | 41 | 789 |
| GO:0008022 | protein C-terminus binding | 1.06e-01 | 1.00e+00 | 3.163 | 1 | 8 | 161 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.08e-01 | 1.00e+00 | 3.137 | 1 | 15 | 164 |
| GO:0034641 | cellular nitrogen compound metabolic process | 1.12e-01 | 1.00e+00 | 3.076 | 1 | 2 | 171 |
| GO:0031625 | ubiquitin protein ligase binding | 1.17e-01 | 1.00e+00 | 3.018 | 1 | 5 | 178 |
| GO:0019904 | protein domain specific binding | 1.17e-01 | 1.00e+00 | 3.010 | 1 | 8 | 179 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.21e-01 | 1.00e+00 | 2.971 | 1 | 22 | 184 |
| GO:0016071 | mRNA metabolic process | 1.44e-01 | 1.00e+00 | 2.693 | 1 | 8 | 223 |
| GO:0008380 | RNA splicing | 1.47e-01 | 1.00e+00 | 2.661 | 1 | 22 | 228 |
| GO:0030425 | dendrite | 1.49e-01 | 1.00e+00 | 2.649 | 1 | 6 | 230 |
| GO:0016070 | RNA metabolic process | 1.59e-01 | 1.00e+00 | 2.546 | 1 | 8 | 247 |
| GO:0006281 | DNA repair | 1.67e-01 | 1.00e+00 | 2.466 | 1 | 14 | 261 |
| GO:0007283 | spermatogenesis | 1.68e-01 | 1.00e+00 | 2.461 | 1 | 3 | 262 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 1.69e-01 | 1.00e+00 | 2.450 | 1 | 23 | 264 |
| GO:0006954 | inflammatory response | 1.76e-01 | 1.00e+00 | 2.380 | 1 | 6 | 277 |
| GO:0005856 | cytoskeleton | 1.84e-01 | 1.00e+00 | 2.309 | 1 | 9 | 291 |
| GO:0043234 | protein complex | 1.87e-01 | 1.00e+00 | 2.290 | 1 | 11 | 295 |
| GO:0030154 | cell differentiation | 1.90e-01 | 1.00e+00 | 2.261 | 1 | 5 | 301 |
| GO:0019901 | protein kinase binding | 1.99e-01 | 1.00e+00 | 2.186 | 1 | 18 | 317 |
| GO:0008283 | cell proliferation | 2.06e-01 | 1.00e+00 | 2.137 | 1 | 14 | 328 |
| GO:0030054 | cell junction | 2.10e-01 | 1.00e+00 | 2.102 | 1 | 5 | 336 |
| GO:0043565 | sequence-specific DNA binding | 2.16e-01 | 1.00e+00 | 2.060 | 1 | 11 | 346 |
| GO:0008285 | negative regulation of cell proliferation | 2.20e-01 | 1.00e+00 | 2.031 | 1 | 10 | 353 |
| GO:0070062 | extracellular vesicular exosome | 2.24e-01 | 1.00e+00 | 0.850 | 3 | 57 | 2400 |
| GO:0000139 | Golgi membrane | 2.24e-01 | 1.00e+00 | 1.998 | 1 | 7 | 361 |
| GO:0008150 | biological_process | 2.39e-01 | 1.00e+00 | 1.891 | 1 | 4 | 389 |
| GO:0009986 | cell surface | 2.46e-01 | 1.00e+00 | 1.843 | 1 | 9 | 402 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.52e-01 | 1.00e+00 | 1.804 | 1 | 24 | 413 |
| GO:0043066 | negative regulation of apoptotic process | 2.58e-01 | 1.00e+00 | 1.766 | 1 | 16 | 424 |
| GO:0006351 | transcription, DNA-templated | 2.65e-01 | 1.00e+00 | 0.996 | 2 | 57 | 1446 |
| GO:0003674 | molecular_function | 2.69e-01 | 1.00e+00 | 1.700 | 1 | 1 | 444 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 2.86e-01 | 1.00e+00 | 1.593 | 1 | 24 | 478 |
| GO:0016032 | viral process | 3.14e-01 | 1.00e+00 | 1.433 | 1 | 26 | 534 |
| GO:0005730 | nucleolus | 3.18e-01 | 1.00e+00 | 0.814 | 2 | 66 | 1641 |
| GO:0016020 | membrane | 3.28e-01 | 1.00e+00 | 0.779 | 2 | 46 | 1681 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.32e-01 | 1.00e+00 | 1.339 | 1 | 37 | 570 |
| GO:0005739 | mitochondrion | 5.12e-01 | 1.00e+00 | 0.531 | 1 | 23 | 998 |
| GO:0008270 | zinc ion binding | 5.12e-01 | 1.00e+00 | 0.533 | 1 | 27 | 997 |
| GO:0044281 | small molecule metabolic process | 5.84e-01 | 1.00e+00 | 0.252 | 1 | 20 | 1211 |
| GO:0005886 | plasma membrane | 8.61e-01 | 1.00e+00 | -0.840 | 1 | 49 | 2582 |