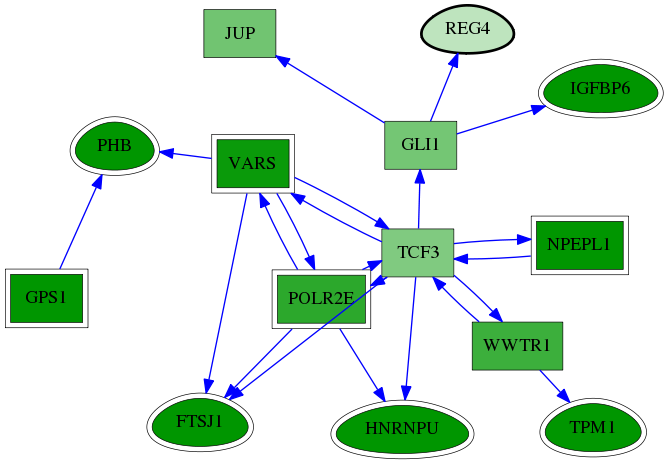
| GO:0001649 | osteoblast differentiation | 2.87e-06 | 2.80e-02 | 5.237 | 4 | 5 | 74 |
| GO:0044212 | transcription regulatory region DNA binding | 9.94e-04 | 1.00e+00 | 3.871 | 3 | 13 | 143 |
| GO:0005667 | transcription factor complex | 1.08e-03 | 1.00e+00 | 3.831 | 3 | 8 | 147 |
| GO:0003065 | positive regulation of heart rate by epinephrine | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0004832 | valine-tRNA ligase activity | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0005199 | structural constituent of cell wall | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0002128 | tRNA nucleoside ribose methylation | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0060390 | regulation of SMAD protein import into nucleus | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:2001065 | mannan binding | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0008175 | tRNA methyltransferase activity | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0006438 | valyl-tRNA aminoacylation | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0071665 | gamma-catenin-TCF7L2 complex | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 1 |
| GO:0003713 | transcription coactivator activity | 2.06e-03 | 1.00e+00 | 3.507 | 3 | 21 | 184 |
| GO:0071603 | endothelial cell-cell adhesion | 2.87e-03 | 1.00e+00 | 8.446 | 1 | 1 | 2 |
| GO:0060032 | notochord regression | 2.87e-03 | 1.00e+00 | 8.446 | 1 | 1 | 2 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 3.12e-03 | 1.00e+00 | 4.563 | 2 | 5 | 59 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.23e-03 | 1.00e+00 | 2.613 | 4 | 29 | 456 |
| GO:0005737 | cytoplasm | 3.77e-03 | 1.00e+00 | 1.253 | 9 | 127 | 2633 |
| GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity | 4.00e-03 | 1.00e+00 | 4.380 | 2 | 5 | 67 |
| GO:0070644 | vitamin D response element binding | 4.30e-03 | 1.00e+00 | 7.861 | 1 | 1 | 3 |
| GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 4.30e-03 | 1.00e+00 | 7.861 | 1 | 1 | 3 |
| GO:0033152 | immunoglobulin V(D)J recombination | 4.30e-03 | 1.00e+00 | 7.861 | 1 | 1 | 3 |
| GO:0003136 | negative regulation of heart induction by canonical Wnt signaling pathway | 4.30e-03 | 1.00e+00 | 7.861 | 1 | 1 | 3 |
| GO:0005862 | muscle thin filament tropomyosin | 4.30e-03 | 1.00e+00 | 7.861 | 1 | 1 | 3 |
| GO:0007418 | ventral midline development | 4.30e-03 | 1.00e+00 | 7.861 | 1 | 1 | 3 |
| GO:0002326 | B cell lineage commitment | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 4 |
| GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 4 |
| GO:0002159 | desmosome assembly | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 4 |
| GO:0010467 | gene expression | 5.74e-03 | 1.00e+00 | 2.383 | 4 | 49 | 535 |
| GO:0005654 | nucleoplasm | 5.78e-03 | 1.00e+00 | 1.993 | 5 | 68 | 876 |
| GO:0009913 | epidermal cell differentiation | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 2 | 5 |
| GO:0071681 | cellular response to indole-3-methanol | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0008235 | metalloexopeptidase activity | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 2 | 5 |
| GO:0021696 | cerebellar cortex morphogenesis | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 2 | 5 |
| GO:0035414 | negative regulation of catenin import into nucleus | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0000788 | nuclear nucleosome | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0086069 | bundle of His cell to Purkinje myocyte communication | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0032059 | bleb | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0002161 | aminoacyl-tRNA editing activity | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 5 |
| GO:0050847 | progesterone receptor signaling pathway | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 2 | 6 |
| GO:0002181 | cytoplasmic translation | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 1 | 6 |
| GO:0001055 | RNA polymerase II activity | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 3 | 6 |
| GO:0005915 | zonula adherens | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 1 | 6 |
| GO:0048599 | oocyte development | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 1 | 6 |
| GO:0017145 | stem cell division | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 2 | 6 |
| GO:0045294 | alpha-catenin binding | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 1 | 6 |
| GO:0050982 | detection of mechanical stimulus | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 1 | 7 |
| GO:0030056 | hemidesmosome | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 1 | 7 |
| GO:0048541 | Peyer's patch development | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 2 | 7 |
| GO:0016342 | catenin complex | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 1 | 7 |
| GO:0031994 | insulin-like growth factor I binding | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 1 | 7 |
| GO:0032835 | glomerulus development | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 1 | 7 |
| GO:0016327 | apicolateral plasma membrane | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 1 | 7 |
| GO:0006450 | regulation of translational fidelity | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 8 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 8 |
| GO:0003181 | atrioventricular valve morphogenesis | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 8 |
| GO:0010388 | cullin deneddylation | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 4 | 8 |
| GO:0071354 | cellular response to interleukin-6 | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 9 |
| GO:0005916 | fascia adherens | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 9 |
| GO:0016331 | morphogenesis of embryonic epithelium | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 9 |
| GO:0034614 | cellular response to reactive oxygen species | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 2 | 9 |
| GO:0006937 | regulation of muscle contraction | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 9 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.40e-02 | 1.00e+00 | 3.446 | 2 | 15 | 128 |
| GO:0005736 | DNA-directed RNA polymerase I complex | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 4 | 10 |
| GO:0005095 | GTPase inhibitor activity | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 10 |
| GO:0031529 | ruffle organization | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 2 | 10 |
| GO:0086005 | ventricular cardiac muscle cell action potential | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 10 |
| GO:0001779 | natural killer cell differentiation | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 10 |
| GO:0001054 | RNA polymerase I activity | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 4 | 10 |
| GO:0048468 | cell development | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 10 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 1.51e-02 | 1.00e+00 | 2.473 | 3 | 21 | 377 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 4 | 11 |
| GO:0035326 | enhancer binding | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 11 |
| GO:0007016 | cytoskeletal anchoring at plasma membrane | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 11 |
| GO:0048546 | digestive tract morphogenesis | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 11 |
| GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 11 |
| GO:0086091 | regulation of heart rate by cardiac conduction | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 11 |
| GO:0007398 | ectoderm development | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 11 |
| GO:0005634 | nucleus | 1.70e-02 | 1.00e+00 | 0.952 | 9 | 158 | 3246 |
| GO:0042307 | positive regulation of protein import into nucleus | 1.71e-02 | 1.00e+00 | 5.861 | 1 | 3 | 12 |
| GO:0090002 | establishment of protein localization to plasma membrane | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 1 | 13 |
| GO:0008589 | regulation of smoothened signaling pathway | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 1 | 13 |
| GO:0005666 | DNA-directed RNA polymerase III complex | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 13 |
| GO:0005665 | DNA-directed RNA polymerase II, core complex | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 13 |
| GO:0001056 | RNA polymerase III activity | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 13 |
| GO:0045214 | sarcomere organization | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 1 | 13 |
| GO:0032993 | protein-DNA complex | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 1 | 13 |
| GO:0006386 | termination of RNA polymerase III transcription | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 3 | 14 |
| GO:0031941 | filamentous actin | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 1 | 14 |
| GO:0006385 | transcription elongation from RNA polymerase III promoter | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 3 | 14 |
| GO:0016575 | histone deacetylation | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 2 | 14 |
| GO:0004177 | aminopeptidase activity | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 3 | 14 |
| GO:0072372 | primary cilium | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 1 | 14 |
| GO:0042803 | protein homodimerization activity | 2.12e-02 | 1.00e+00 | 2.290 | 3 | 13 | 428 |
| GO:0030057 | desmosome | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 2 | 15 |
| GO:0032781 | positive regulation of ATPase activity | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 2 | 15 |
| GO:0000188 | inactivation of MAPK activity | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 1 | 15 |
| GO:0045880 | positive regulation of smoothened signaling pathway | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 1 | 15 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 2.16e-02 | 1.00e+00 | 3.115 | 2 | 18 | 161 |
| GO:0008380 | RNA splicing | 2.26e-02 | 1.00e+00 | 3.080 | 2 | 21 | 165 |
| GO:0009954 | proximal/distal pattern formation | 2.41e-02 | 1.00e+00 | 5.359 | 1 | 2 | 17 |
| GO:0007369 | gastrulation | 2.41e-02 | 1.00e+00 | 5.359 | 1 | 2 | 17 |
| GO:0009953 | dorsal/ventral pattern formation | 2.55e-02 | 1.00e+00 | 5.276 | 1 | 2 | 18 |
| GO:0003899 | DNA-directed RNA polymerase activity | 2.55e-02 | 1.00e+00 | 5.276 | 1 | 3 | 18 |
| GO:0030017 | sarcomere | 2.55e-02 | 1.00e+00 | 5.276 | 1 | 2 | 18 |
| GO:0031435 | mitogen-activated protein kinase kinase kinase binding | 2.55e-02 | 1.00e+00 | 5.276 | 1 | 1 | 18 |
| GO:0043588 | skin development | 2.69e-02 | 1.00e+00 | 5.198 | 1 | 1 | 19 |
| GO:0035329 | hippo signaling | 2.69e-02 | 1.00e+00 | 5.198 | 1 | 1 | 19 |
| GO:0030049 | muscle filament sliding | 2.69e-02 | 1.00e+00 | 5.198 | 1 | 2 | 19 |
| GO:0045787 | positive regulation of cell cycle | 2.69e-02 | 1.00e+00 | 5.198 | 1 | 1 | 19 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 2.69e-02 | 1.00e+00 | 5.198 | 1 | 1 | 19 |
| GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 1 | 20 |
| GO:0045296 | cadherin binding | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 2 | 20 |
| GO:0010718 | positive regulation of epithelial to mesenchymal transition | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 2 | 20 |
| GO:0071897 | DNA biosynthetic process | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 2 | 20 |
| GO:0043425 | bHLH transcription factor binding | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 1 | 20 |
| GO:0005829 | cytosol | 2.93e-02 | 1.00e+00 | 1.228 | 6 | 88 | 1787 |
| GO:0005856 | cytoskeleton | 2.94e-02 | 1.00e+00 | 2.876 | 2 | 8 | 190 |
| GO:0008307 | structural constituent of muscle | 3.11e-02 | 1.00e+00 | 4.987 | 1 | 2 | 22 |
| GO:0051149 | positive regulation of muscle cell differentiation | 3.11e-02 | 1.00e+00 | 4.987 | 1 | 2 | 22 |
| GO:0021983 | pituitary gland development | 3.11e-02 | 1.00e+00 | 4.987 | 1 | 2 | 22 |
| GO:0043967 | histone H4 acetylation | 3.11e-02 | 1.00e+00 | 4.987 | 1 | 1 | 22 |
| GO:0014704 | intercalated disc | 3.25e-02 | 1.00e+00 | 4.922 | 1 | 2 | 23 |
| GO:0070491 | repressing transcription factor binding | 3.25e-02 | 1.00e+00 | 4.922 | 1 | 2 | 23 |
| GO:0006370 | 7-methylguanosine mRNA capping | 3.39e-02 | 1.00e+00 | 4.861 | 1 | 5 | 24 |
| GO:0008180 | COP9 signalosome | 3.39e-02 | 1.00e+00 | 4.861 | 1 | 6 | 24 |
| GO:0008016 | regulation of heart contraction | 3.39e-02 | 1.00e+00 | 4.861 | 1 | 1 | 24 |
| GO:0070888 | E-box binding | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 1 | 25 |
| GO:0005913 | cell-cell adherens junction | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 3 | 25 |
| GO:0060271 | cilium morphogenesis | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 1 | 26 |
| GO:0030145 | manganese ion binding | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 3 | 26 |
| GO:0051496 | positive regulation of stress fiber assembly | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 2 | 26 |
| GO:0033077 | T cell differentiation in thymus | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 2 | 26 |
| GO:0060048 | cardiac muscle contraction | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 1 | 26 |
| GO:0006360 | transcription from RNA polymerase I promoter | 3.80e-02 | 1.00e+00 | 4.691 | 1 | 4 | 27 |
| GO:0034332 | adherens junction organization | 3.94e-02 | 1.00e+00 | 4.639 | 1 | 1 | 28 |
| GO:0009898 | cytoplasmic side of plasma membrane | 3.94e-02 | 1.00e+00 | 4.639 | 1 | 3 | 28 |
| GO:0043966 | histone H3 acetylation | 3.94e-02 | 1.00e+00 | 4.639 | 1 | 4 | 28 |
| GO:0042692 | muscle cell differentiation | 3.94e-02 | 1.00e+00 | 4.639 | 1 | 2 | 28 |
| GO:0003677 | DNA binding | 3.99e-02 | 1.00e+00 | 1.559 | 4 | 52 | 947 |
| GO:0045599 | negative regulation of fat cell differentiation | 4.08e-02 | 1.00e+00 | 4.588 | 1 | 2 | 29 |
| GO:0006418 | tRNA aminoacylation for protein translation | 4.08e-02 | 1.00e+00 | 4.588 | 1 | 4 | 29 |
| GO:0045785 | positive regulation of cell adhesion | 4.08e-02 | 1.00e+00 | 4.588 | 1 | 3 | 29 |
| GO:0006383 | transcription from RNA polymerase III promoter | 4.08e-02 | 1.00e+00 | 4.588 | 1 | 6 | 29 |
| GO:0001933 | negative regulation of protein phosphorylation | 4.22e-02 | 1.00e+00 | 4.539 | 1 | 3 | 30 |
| GO:0016328 | lateral plasma membrane | 4.36e-02 | 1.00e+00 | 4.492 | 1 | 2 | 31 |
| GO:0045740 | positive regulation of DNA replication | 4.49e-02 | 1.00e+00 | 4.446 | 1 | 1 | 32 |
| GO:0030890 | positive regulation of B cell proliferation | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 2 | 33 |
| GO:0007254 | JNK cascade | 4.77e-02 | 1.00e+00 | 4.359 | 1 | 1 | 34 |
| GO:0000902 | cell morphogenesis | 4.91e-02 | 1.00e+00 | 4.317 | 1 | 5 | 35 |
| GO:0050434 | positive regulation of viral transcription | 4.91e-02 | 1.00e+00 | 4.317 | 1 | 5 | 35 |
| GO:0003682 | chromatin binding | 4.92e-02 | 1.00e+00 | 2.469 | 2 | 15 | 252 |
| GO:0008092 | cytoskeletal protein binding | 5.04e-02 | 1.00e+00 | 4.276 | 1 | 1 | 36 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 5.04e-02 | 1.00e+00 | 4.276 | 1 | 2 | 36 |
| GO:0007224 | smoothened signaling pathway | 5.45e-02 | 1.00e+00 | 4.161 | 1 | 1 | 39 |
| GO:0006396 | RNA processing | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 4 | 40 |
| GO:0006283 | transcription-coupled nucleotide-excision repair | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 6 | 40 |
| GO:0005882 | intermediate filament | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 1 | 40 |
| GO:0008285 | negative regulation of cell proliferation | 5.83e-02 | 1.00e+00 | 2.332 | 2 | 11 | 277 |
| GO:0008284 | positive regulation of cell proliferation | 5.90e-02 | 1.00e+00 | 2.322 | 2 | 8 | 279 |
| GO:0030183 | B cell differentiation | 6.00e-02 | 1.00e+00 | 4.020 | 1 | 1 | 43 |
| GO:0051291 | protein heterooligomerization | 6.00e-02 | 1.00e+00 | 4.020 | 1 | 1 | 43 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 6.02e-02 | 1.00e+00 | 1.700 | 3 | 29 | 644 |
| GO:0045216 | cell-cell junction organization | 6.13e-02 | 1.00e+00 | 3.987 | 1 | 1 | 44 |
| GO:0001725 | stress fiber | 6.40e-02 | 1.00e+00 | 3.922 | 1 | 6 | 46 |
| GO:0019903 | protein phosphatase binding | 6.54e-02 | 1.00e+00 | 3.891 | 1 | 2 | 47 |
| GO:0032481 | positive regulation of type I interferon production | 6.67e-02 | 1.00e+00 | 3.861 | 1 | 3 | 48 |
| GO:0001558 | regulation of cell growth | 6.80e-02 | 1.00e+00 | 3.831 | 1 | 2 | 49 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 6.94e-02 | 1.00e+00 | 3.802 | 1 | 2 | 50 |
| GO:0032587 | ruffle membrane | 6.94e-02 | 1.00e+00 | 3.802 | 1 | 3 | 50 |
| GO:0006469 | negative regulation of protein kinase activity | 6.94e-02 | 1.00e+00 | 3.802 | 1 | 2 | 50 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 7.21e-02 | 1.00e+00 | 3.746 | 1 | 8 | 52 |
| GO:0006289 | nucleotide-excision repair | 7.34e-02 | 1.00e+00 | 3.718 | 1 | 6 | 53 |
| GO:0045666 | positive regulation of neuron differentiation | 7.34e-02 | 1.00e+00 | 3.718 | 1 | 3 | 53 |
| GO:0034329 | cell junction assembly | 7.34e-02 | 1.00e+00 | 3.718 | 1 | 2 | 53 |
| GO:0006936 | muscle contraction | 7.47e-02 | 1.00e+00 | 3.691 | 1 | 5 | 54 |
| GO:0042060 | wound healing | 7.61e-02 | 1.00e+00 | 3.665 | 1 | 1 | 55 |
| GO:0006355 | regulation of transcription, DNA-templated | 7.76e-02 | 1.00e+00 | 1.549 | 3 | 31 | 715 |
| GO:0003705 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity | 8.01e-02 | 1.00e+00 | 3.588 | 1 | 3 | 58 |
| GO:0042826 | histone deacetylase binding | 8.01e-02 | 1.00e+00 | 3.588 | 1 | 5 | 58 |
| GO:0030336 | negative regulation of cell migration | 8.27e-02 | 1.00e+00 | 3.539 | 1 | 3 | 60 |
| GO:0006366 | transcription from RNA polymerase II promoter | 8.39e-02 | 1.00e+00 | 2.032 | 2 | 23 | 341 |
| GO:0071013 | catalytic step 2 spliceosome | 8.40e-02 | 1.00e+00 | 3.515 | 1 | 5 | 61 |
| GO:0007010 | cytoskeleton organization | 8.40e-02 | 1.00e+00 | 3.515 | 1 | 4 | 61 |
| GO:0016337 | single organismal cell-cell adhesion | 8.54e-02 | 1.00e+00 | 3.492 | 1 | 3 | 62 |
| GO:0030324 | lung development | 8.54e-02 | 1.00e+00 | 3.492 | 1 | 1 | 62 |
| GO:0050821 | protein stabilization | 8.80e-02 | 1.00e+00 | 3.446 | 1 | 3 | 64 |
| GO:0030018 | Z disc | 8.80e-02 | 1.00e+00 | 3.446 | 1 | 3 | 64 |
| GO:0005200 | structural constituent of cytoskeleton | 9.33e-02 | 1.00e+00 | 3.359 | 1 | 2 | 68 |
| GO:0006928 | cellular component movement | 9.59e-02 | 1.00e+00 | 3.317 | 1 | 6 | 70 |
| GO:0030308 | negative regulation of cell growth | 9.59e-02 | 1.00e+00 | 3.317 | 1 | 3 | 70 |
| GO:0042127 | regulation of cell proliferation | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 4 | 80 |
| GO:0030529 | ribonucleoprotein complex | 1.11e-01 | 1.00e+00 | 3.088 | 1 | 5 | 82 |
| GO:0008201 | heparin binding | 1.23e-01 | 1.00e+00 | 2.938 | 1 | 4 | 91 |
| GO:0032496 | response to lipopolysaccharide | 1.25e-01 | 1.00e+00 | 2.907 | 1 | 1 | 93 |
| GO:0016477 | cell migration | 1.29e-01 | 1.00e+00 | 2.861 | 1 | 3 | 96 |
| GO:0008017 | microtubule binding | 1.31e-01 | 1.00e+00 | 2.846 | 1 | 9 | 97 |
| GO:0005198 | structural molecule activity | 1.36e-01 | 1.00e+00 | 2.788 | 1 | 4 | 101 |
| GO:0005911 | cell-cell junction | 1.39e-01 | 1.00e+00 | 2.746 | 1 | 3 | 104 |
| GO:0016323 | basolateral plasma membrane | 1.42e-01 | 1.00e+00 | 2.718 | 1 | 3 | 106 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 1.43e-01 | 1.00e+00 | 2.705 | 1 | 5 | 107 |
| GO:0000790 | nuclear chromatin | 1.46e-01 | 1.00e+00 | 2.678 | 1 | 9 | 109 |
| GO:0042981 | regulation of apoptotic process | 1.49e-01 | 1.00e+00 | 2.639 | 1 | 4 | 112 |
| GO:0007049 | cell cycle | 1.53e-01 | 1.00e+00 | 2.601 | 1 | 8 | 115 |
| GO:0015629 | actin cytoskeleton | 1.78e-01 | 1.00e+00 | 2.359 | 1 | 5 | 136 |
| GO:0003714 | transcription corepressor activity | 1.80e-01 | 1.00e+00 | 2.348 | 1 | 5 | 137 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.86e-01 | 1.00e+00 | 2.296 | 1 | 7 | 142 |
| GO:0005515 | protein binding | 1.94e-01 | 1.00e+00 | 0.436 | 8 | 198 | 4124 |
| GO:0007399 | nervous system development | 1.97e-01 | 1.00e+00 | 2.198 | 1 | 4 | 152 |
| GO:0001701 | in utero embryonic development | 2.00e-01 | 1.00e+00 | 2.179 | 1 | 7 | 154 |
| GO:0007283 | spermatogenesis | 2.16e-01 | 1.00e+00 | 2.054 | 1 | 4 | 168 |
| GO:0007165 | signal transduction | 2.21e-01 | 1.00e+00 | 1.175 | 2 | 24 | 618 |
| GO:0003779 | actin binding | 2.28e-01 | 1.00e+00 | 1.962 | 1 | 3 | 179 |
| GO:0005739 | mitochondrion | 2.43e-01 | 1.00e+00 | 1.082 | 2 | 32 | 659 |
| GO:0005576 | extracellular region | 2.45e-01 | 1.00e+00 | 1.073 | 2 | 15 | 663 |
| GO:0005743 | mitochondrial inner membrane | 2.48e-01 | 1.00e+00 | 1.824 | 1 | 9 | 197 |
| GO:0006281 | DNA repair | 2.55e-01 | 1.00e+00 | 1.781 | 1 | 24 | 203 |
| GO:0008134 | transcription factor binding | 2.61e-01 | 1.00e+00 | 1.739 | 1 | 10 | 209 |
| GO:0043234 | protein complex | 2.63e-01 | 1.00e+00 | 1.732 | 1 | 13 | 210 |
| GO:0019899 | enzyme binding | 2.63e-01 | 1.00e+00 | 1.732 | 1 | 7 | 210 |
| GO:0043231 | intracellular membrane-bounded organelle | 2.71e-01 | 1.00e+00 | 1.678 | 1 | 8 | 218 |
| GO:0042493 | response to drug | 2.75e-01 | 1.00e+00 | 1.652 | 1 | 4 | 222 |
| GO:0019901 | protein kinase binding | 2.86e-01 | 1.00e+00 | 1.588 | 1 | 8 | 232 |
| GO:0003723 | RNA binding | 3.02e-01 | 1.00e+00 | 1.498 | 1 | 22 | 247 |
| GO:0006508 | proteolysis | 3.19e-01 | 1.00e+00 | 1.402 | 1 | 11 | 264 |
| GO:0046982 | protein heterodimerization activity | 3.20e-01 | 1.00e+00 | 1.396 | 1 | 11 | 265 |
| GO:0043565 | sequence-specific DNA binding | 3.30e-01 | 1.00e+00 | 1.343 | 1 | 8 | 275 |
| GO:0005925 | focal adhesion | 3.37e-01 | 1.00e+00 | 1.306 | 1 | 14 | 282 |
| GO:0009986 | cell surface | 3.45e-01 | 1.00e+00 | 1.261 | 1 | 5 | 291 |
| GO:0005524 | ATP binding | 3.70e-01 | 1.00e+00 | 0.645 | 2 | 37 | 892 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.70e-01 | 1.00e+00 | 1.138 | 1 | 17 | 317 |
| GO:0005509 | calcium ion binding | 4.01e-01 | 1.00e+00 | 0.991 | 1 | 9 | 351 |
| GO:0044267 | cellular protein metabolic process | 4.02e-01 | 1.00e+00 | 0.987 | 1 | 14 | 352 |
| GO:0070062 | extracellular vesicular exosome | 4.26e-01 | 1.00e+00 | 0.351 | 3 | 51 | 1641 |
| GO:0005794 | Golgi apparatus | 4.56e-01 | 1.00e+00 | 0.749 | 1 | 6 | 415 |
| GO:0016032 | viral process | 4.66e-01 | 1.00e+00 | 0.705 | 1 | 30 | 428 |
| GO:0006351 | transcription, DNA-templated | 4.67e-01 | 1.00e+00 | 0.375 | 2 | 47 | 1076 |
| GO:0045087 | innate immune response | 4.85e-01 | 1.00e+00 | 0.626 | 1 | 12 | 452 |
| GO:0016020 | membrane | 5.31e-01 | 1.00e+00 | 0.209 | 2 | 56 | 1207 |
| GO:0005730 | nucleolus | 5.36e-01 | 1.00e+00 | 0.197 | 2 | 74 | 1217 |
| GO:0005887 | integral component of plasma membrane | 5.73e-01 | 1.00e+00 | 0.279 | 1 | 8 | 575 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 5.77e-01 | 1.00e+00 | 0.261 | 1 | 22 | 582 |
| GO:0005615 | extracellular space | 6.20e-01 | 1.00e+00 | 0.100 | 1 | 15 | 651 |
| GO:0044822 | poly(A) RNA binding | 6.98e-01 | 1.00e+00 | -0.196 | 1 | 45 | 799 |
| GO:0046872 | metal ion binding | 7.55e-01 | 1.00e+00 | -0.421 | 1 | 24 | 934 |
| GO:0005886 | plasma membrane | 9.41e-01 | 1.00e+00 | -1.355 | 1 | 46 | 1784 |