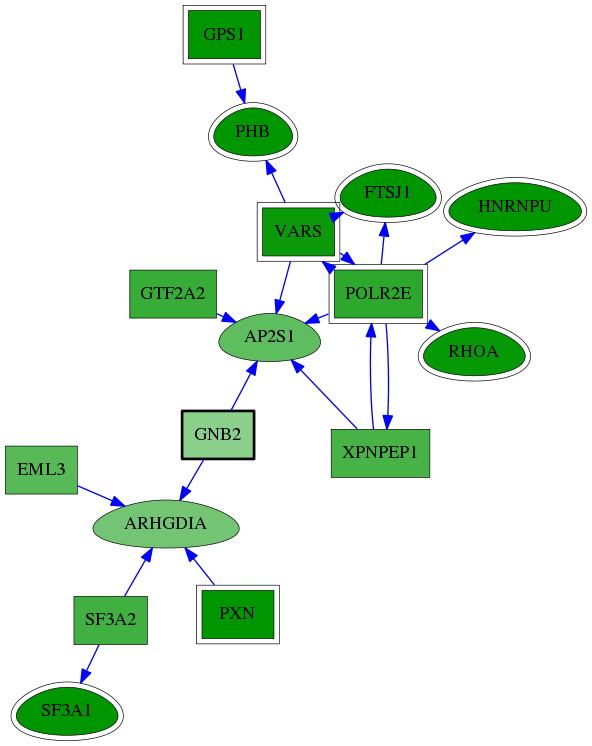
| GO:0000389 | mRNA 3'-splice site recognition | 1.51e-05 | 1.47e-01 | 8.253 | 2 | 2 | 4 |
| GO:0000398 | mRNA splicing, via spliceosome | 4.54e-05 | 4.43e-01 | 4.253 | 4 | 15 | 128 |
| GO:0008380 | RNA splicing | 1.22e-04 | 1.00e+00 | 3.887 | 4 | 21 | 165 |
| GO:0071013 | catalytic step 2 spliceosome | 1.23e-04 | 1.00e+00 | 4.908 | 3 | 5 | 61 |
| GO:0010467 | gene expression | 1.31e-04 | 1.00e+00 | 2.775 | 6 | 49 | 535 |
| GO:0050772 | positive regulation of axonogenesis | 1.94e-04 | 1.00e+00 | 6.553 | 2 | 2 | 13 |
| GO:0050770 | regulation of axonogenesis | 1.94e-04 | 1.00e+00 | 6.553 | 2 | 3 | 13 |
| GO:0050771 | negative regulation of axonogenesis | 2.26e-04 | 1.00e+00 | 6.446 | 2 | 2 | 14 |
| GO:0007266 | Rho protein signal transduction | 1.45e-03 | 1.00e+00 | 5.124 | 2 | 2 | 35 |
| GO:0004832 | valine-tRNA ligase activity | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 1 |
| GO:0061383 | trabecula morphogenesis | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 1 |
| GO:0002128 | tRNA nucleoside ribose methylation | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 1 |
| GO:0008175 | tRNA methyltransferase activity | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 1 |
| GO:0051435 | BH4 domain binding | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 1 |
| GO:0006438 | valyl-tRNA aminoacylation | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 1 |
| GO:0005654 | nucleoplasm | 1.87e-03 | 1.00e+00 | 2.064 | 6 | 68 | 876 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 3.18e-03 | 1.00e+00 | 4.553 | 2 | 8 | 52 |
| GO:0005684 | U2-type spliceosomal complex | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 2 | 2 |
| GO:0033688 | regulation of osteoblast proliferation | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 2 |
| GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 2 |
| GO:0005681 | spliceosomal complex | 3.55e-03 | 1.00e+00 | 4.472 | 2 | 8 | 55 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 3.84e-03 | 1.00e+00 | 3.202 | 3 | 8 | 199 |
| GO:0016032 | viral process | 4.35e-03 | 1.00e+00 | 2.512 | 4 | 30 | 428 |
| GO:0005094 | Rho GDP-dissociation inhibitor activity | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 3 |
| GO:0070006 | metalloaminopeptidase activity | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 3 |
| GO:0006928 | cellular component movement | 5.70e-03 | 1.00e+00 | 4.124 | 2 | 6 | 70 |
| GO:0001649 | osteoblast differentiation | 6.35e-03 | 1.00e+00 | 4.044 | 2 | 5 | 74 |
| GO:0036089 | cleavage furrow formation | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 1 | 4 |
| GO:0010815 | bradykinin catabolic process | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 1 | 4 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 6.68e-03 | 1.00e+00 | 4.005 | 2 | 2 | 76 |
| GO:0005938 | cell cortex | 6.86e-03 | 1.00e+00 | 3.987 | 2 | 6 | 77 |
| GO:0038027 | apolipoprotein A-I-mediated signaling pathway | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 2 | 5 |
| GO:0043931 | ossification involved in bone maturation | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 5 |
| GO:0005672 | transcription factor TFIIA complex | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 5 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 5 |
| GO:0002161 | aminoacyl-tRNA editing activity | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 5 |
| GO:0030027 | lamellipodium | 8.88e-03 | 1.00e+00 | 3.794 | 2 | 3 | 88 |
| GO:0050847 | progesterone receptor signaling pathway | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 2 | 6 |
| GO:0002181 | cytoplasmic translation | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 1 | 6 |
| GO:0001055 | RNA polymerase II activity | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 3 | 6 |
| GO:0043297 | apical junction assembly | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 2 | 6 |
| GO:0005925 | focal adhesion | 1.01e-02 | 1.00e+00 | 2.699 | 3 | 14 | 282 |
| GO:0051020 | GTPase binding | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 7 |
| GO:0035615 | clathrin adaptor activity | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 7 |
| GO:0007172 | signal complex assembly | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 7 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 1.29e-02 | 1.00e+00 | 3.512 | 2 | 5 | 107 |
| GO:0090307 | spindle assembly involved in mitosis | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 2 | 8 |
| GO:0006450 | regulation of translational fidelity | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 1 | 8 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 1 | 8 |
| GO:0010388 | cullin deneddylation | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 4 | 8 |
| GO:0060396 | growth hormone receptor signaling pathway | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 1 | 8 |
| GO:0043296 | apical junction complex | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 1 | 8 |
| GO:0006397 | mRNA processing | 1.36e-02 | 1.00e+00 | 3.472 | 2 | 12 | 110 |
| GO:0030122 | AP-2 adaptor complex | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 2 | 9 |
| GO:0071354 | cellular response to interleukin-6 | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 1 | 9 |
| GO:0030100 | regulation of endocytosis | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 1 | 9 |
| GO:0048268 | clathrin coat assembly | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 2 | 9 |
| GO:0034614 | cellular response to reactive oxygen species | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 2 | 9 |
| GO:0072583 | clathrin-mediated endocytosis | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 1 | 9 |
| GO:0003924 | GTPase activity | 1.58e-02 | 1.00e+00 | 3.359 | 2 | 7 | 119 |
| GO:0005736 | DNA-directed RNA polymerase I complex | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 4 | 10 |
| GO:0005095 | GTPase inhibitor activity | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 1 | 10 |
| GO:0050919 | negative chemotaxis | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 1 | 10 |
| GO:0005246 | calcium channel regulator activity | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 1 | 10 |
| GO:0017166 | vinculin binding | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 1 | 10 |
| GO:0001054 | RNA polymerase I activity | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 4 | 10 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 4 | 11 |
| GO:0006184 | GTP catabolic process | 1.87e-02 | 1.00e+00 | 3.231 | 2 | 7 | 130 |
| GO:0017025 | TBP-class protein binding | 1.95e-02 | 1.00e+00 | 5.668 | 1 | 1 | 12 |
| GO:0005666 | DNA-directed RNA polymerase III complex | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 3 | 13 |
| GO:0005665 | DNA-directed RNA polymerase II, core complex | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 3 | 13 |
| GO:0001056 | RNA polymerase III activity | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 3 | 13 |
| GO:0042346 | positive regulation of NF-kappaB import into nucleus | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 1 | 13 |
| GO:0071526 | semaphorin-plexin signaling pathway | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 1 | 13 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 2.21e-02 | 1.00e+00 | 3.104 | 2 | 5 | 142 |
| GO:0006386 | termination of RNA polymerase III transcription | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 3 | 14 |
| GO:0006385 | transcription elongation from RNA polymerase III promoter | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 3 | 14 |
| GO:0016575 | histone deacetylation | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 2 | 14 |
| GO:0004177 | aminopeptidase activity | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 3 | 14 |
| GO:0000188 | inactivation of MAPK activity | 2.43e-02 | 1.00e+00 | 5.347 | 1 | 1 | 15 |
| GO:0032467 | positive regulation of cytokinesis | 2.59e-02 | 1.00e+00 | 5.253 | 1 | 1 | 16 |
| GO:0017022 | myosin binding | 2.59e-02 | 1.00e+00 | 5.253 | 1 | 2 | 16 |
| GO:0030532 | small nuclear ribonucleoprotein complex | 2.75e-02 | 1.00e+00 | 5.166 | 1 | 1 | 17 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 2.79e-02 | 1.00e+00 | 2.922 | 2 | 18 | 161 |
| GO:0003899 | DNA-directed RNA polymerase activity | 2.91e-02 | 1.00e+00 | 5.083 | 1 | 3 | 18 |
| GO:0005515 | protein binding | 2.96e-02 | 1.00e+00 | 0.703 | 11 | 198 | 4124 |
| GO:0048041 | focal adhesion assembly | 3.07e-02 | 1.00e+00 | 5.005 | 1 | 1 | 19 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 3.07e-02 | 1.00e+00 | 5.005 | 1 | 1 | 19 |
| GO:0007264 | small GTPase mediated signal transduction | 3.22e-02 | 1.00e+00 | 2.810 | 2 | 8 | 174 |
| GO:0030032 | lamellipodium assembly | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 2 | 20 |
| GO:0071377 | cellular response to glucagon stimulus | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 2 | 20 |
| GO:0071897 | DNA biosynthetic process | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 2 | 20 |
| GO:0050690 | regulation of defense response to virus by virus | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 1 | 22 |
| GO:0005875 | microtubule associated complex | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 1 | 22 |
| GO:0048754 | branching morphogenesis of an epithelial tube | 3.71e-02 | 1.00e+00 | 4.730 | 1 | 1 | 23 |
| GO:0001103 | RNA polymerase II repressing transcription factor binding | 3.71e-02 | 1.00e+00 | 4.730 | 1 | 2 | 23 |
| GO:0032880 | regulation of protein localization | 3.71e-02 | 1.00e+00 | 4.730 | 1 | 1 | 23 |
| GO:0001772 | immunological synapse | 3.71e-02 | 1.00e+00 | 4.730 | 1 | 2 | 23 |
| GO:0030669 | clathrin-coated endocytic vesicle membrane | 3.71e-02 | 1.00e+00 | 4.730 | 1 | 1 | 23 |
| GO:0005856 | cytoskeleton | 3.78e-02 | 1.00e+00 | 2.684 | 2 | 8 | 190 |
| GO:0007162 | negative regulation of cell adhesion | 3.86e-02 | 1.00e+00 | 4.668 | 1 | 2 | 24 |
| GO:0006370 | 7-methylguanosine mRNA capping | 3.86e-02 | 1.00e+00 | 4.668 | 1 | 5 | 24 |
| GO:0008180 | COP9 signalosome | 3.86e-02 | 1.00e+00 | 4.668 | 1 | 6 | 24 |
| GO:0034446 | substrate adhesion-dependent cell spreading | 4.02e-02 | 1.00e+00 | 4.610 | 1 | 2 | 25 |
| GO:0030145 | manganese ion binding | 4.18e-02 | 1.00e+00 | 4.553 | 1 | 3 | 26 |
| GO:0051496 | positive regulation of stress fiber assembly | 4.18e-02 | 1.00e+00 | 4.553 | 1 | 2 | 26 |
| GO:0006360 | transcription from RNA polymerase I promoter | 4.34e-02 | 1.00e+00 | 4.499 | 1 | 4 | 27 |
| GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 4.49e-02 | 1.00e+00 | 4.446 | 1 | 2 | 28 |
| GO:0006418 | tRNA aminoacylation for protein translation | 4.65e-02 | 1.00e+00 | 4.395 | 1 | 4 | 29 |
| GO:0006383 | transcription from RNA polymerase III promoter | 4.65e-02 | 1.00e+00 | 4.395 | 1 | 6 | 29 |
| GO:0007268 | synaptic transmission | 4.69e-02 | 1.00e+00 | 2.512 | 2 | 7 | 214 |
| GO:0032154 | cleavage furrow | 4.81e-02 | 1.00e+00 | 4.347 | 1 | 3 | 30 |
| GO:0030054 | cell junction | 4.85e-02 | 1.00e+00 | 2.485 | 2 | 5 | 218 |
| GO:0021762 | substantia nigra development | 5.43e-02 | 1.00e+00 | 4.166 | 1 | 1 | 34 |
| GO:0007254 | JNK cascade | 5.43e-02 | 1.00e+00 | 4.166 | 1 | 1 | 34 |
| GO:0008565 | protein transporter activity | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 2 | 35 |
| GO:0050434 | positive regulation of viral transcription | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 5 | 35 |
| GO:0044297 | cell body | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 5 | 35 |
| GO:0007411 | axon guidance | 5.63e-02 | 1.00e+00 | 2.365 | 2 | 8 | 237 |
| GO:0005829 | cytosol | 5.64e-02 | 1.00e+00 | 1.035 | 6 | 88 | 1787 |
| GO:0030334 | regulation of cell migration | 5.74e-02 | 1.00e+00 | 4.083 | 1 | 2 | 36 |
| GO:0030666 | endocytic vesicle membrane | 5.74e-02 | 1.00e+00 | 4.083 | 1 | 1 | 36 |
| GO:0003723 | RNA binding | 6.06e-02 | 1.00e+00 | 2.305 | 2 | 22 | 247 |
| GO:0006396 | RNA processing | 6.36e-02 | 1.00e+00 | 3.931 | 1 | 4 | 40 |
| GO:0006283 | transcription-coupled nucleotide-excision repair | 6.36e-02 | 1.00e+00 | 3.931 | 1 | 6 | 40 |
| GO:0008013 | beta-catenin binding | 7.13e-02 | 1.00e+00 | 3.762 | 1 | 3 | 45 |
| GO:0001725 | stress fiber | 7.28e-02 | 1.00e+00 | 3.730 | 1 | 6 | 46 |
| GO:0032481 | positive regulation of type I interferon production | 7.59e-02 | 1.00e+00 | 3.668 | 1 | 3 | 48 |
| GO:0006289 | nucleotide-excision repair | 8.35e-02 | 1.00e+00 | 3.525 | 1 | 6 | 53 |
| GO:0045666 | positive regulation of neuron differentiation | 8.35e-02 | 1.00e+00 | 3.525 | 1 | 3 | 53 |
| GO:0034329 | cell junction assembly | 8.35e-02 | 1.00e+00 | 3.525 | 1 | 2 | 53 |
| GO:0006936 | muscle contraction | 8.50e-02 | 1.00e+00 | 3.499 | 1 | 5 | 54 |
| GO:0042826 | histone deacetylase binding | 9.10e-02 | 1.00e+00 | 3.395 | 1 | 5 | 58 |
| GO:0030336 | negative regulation of cell migration | 9.40e-02 | 1.00e+00 | 3.347 | 1 | 3 | 60 |
| GO:0007010 | cytoskeleton organization | 9.55e-02 | 1.00e+00 | 3.323 | 1 | 4 | 61 |
| GO:0000187 | activation of MAPK activity | 9.85e-02 | 1.00e+00 | 3.276 | 1 | 3 | 63 |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 9.99e-02 | 1.00e+00 | 3.253 | 1 | 3 | 64 |
| GO:0006112 | energy reserve metabolic process | 1.01e-01 | 1.00e+00 | 3.231 | 1 | 3 | 65 |
| GO:0007229 | integrin-mediated signaling pathway | 1.03e-01 | 1.00e+00 | 3.209 | 1 | 1 | 66 |
| GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity | 1.04e-01 | 1.00e+00 | 3.187 | 1 | 5 | 67 |
| GO:0005178 | integrin binding | 1.06e-01 | 1.00e+00 | 3.166 | 1 | 3 | 68 |
| GO:0005096 | GTPase activator activity | 1.06e-01 | 1.00e+00 | 3.166 | 1 | 2 | 68 |
| GO:0006366 | transcription from RNA polymerase II promoter | 1.06e-01 | 1.00e+00 | 1.840 | 2 | 23 | 341 |
| GO:0008360 | regulation of cell shape | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 3 | 70 |
| GO:0030308 | negative regulation of cell growth | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 3 | 70 |
| GO:0030496 | midbody | 1.13e-01 | 1.00e+00 | 3.064 | 1 | 6 | 73 |
| GO:0070062 | extracellular vesicular exosome | 1.16e-01 | 1.00e+00 | 0.895 | 5 | 51 | 1641 |
| GO:0030529 | ribonucleoprotein complex | 1.26e-01 | 1.00e+00 | 2.896 | 1 | 5 | 82 |
| GO:0005215 | transporter activity | 1.26e-01 | 1.00e+00 | 2.896 | 1 | 5 | 82 |
| GO:0048015 | phosphatidylinositol-mediated signaling | 1.28e-01 | 1.00e+00 | 2.878 | 1 | 2 | 83 |
| GO:0031982 | vesicle | 1.34e-01 | 1.00e+00 | 2.810 | 1 | 2 | 87 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | 1.36e-01 | 1.00e+00 | 2.778 | 1 | 2 | 89 |
| GO:0006886 | intracellular protein transport | 1.36e-01 | 1.00e+00 | 2.778 | 1 | 2 | 89 |
| GO:0030036 | actin cytoskeleton organization | 1.38e-01 | 1.00e+00 | 2.762 | 1 | 4 | 90 |
| GO:0044822 | poly(A) RNA binding | 1.38e-01 | 1.00e+00 | 1.196 | 3 | 45 | 799 |
| GO:0042803 | protein homodimerization activity | 1.54e-01 | 1.00e+00 | 1.512 | 2 | 13 | 428 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 1.63e-01 | 1.00e+00 | 2.499 | 1 | 2 | 108 |
| GO:0043547 | positive regulation of GTPase activity | 1.69e-01 | 1.00e+00 | 2.446 | 1 | 2 | 112 |
| GO:0042981 | regulation of apoptotic process | 1.69e-01 | 1.00e+00 | 2.446 | 1 | 4 | 112 |
| GO:0007049 | cell cycle | 1.73e-01 | 1.00e+00 | 2.408 | 1 | 8 | 115 |
| GO:0005765 | lysosomal membrane | 1.80e-01 | 1.00e+00 | 2.347 | 1 | 1 | 120 |
| GO:0032403 | protein complex binding | 1.93e-01 | 1.00e+00 | 2.231 | 1 | 5 | 130 |
| GO:0030168 | platelet activation | 2.08e-01 | 1.00e+00 | 2.114 | 1 | 5 | 141 |
| GO:0044212 | transcription regulatory region DNA binding | 2.10e-01 | 1.00e+00 | 2.094 | 1 | 13 | 143 |
| GO:0004871 | signal transducer activity | 2.17e-01 | 1.00e+00 | 2.044 | 1 | 4 | 148 |
| GO:0005874 | microtubule | 2.36e-01 | 1.00e+00 | 1.905 | 1 | 10 | 163 |
| GO:0005737 | cytoplasm | 2.45e-01 | 1.00e+00 | 0.476 | 6 | 127 | 2633 |
| GO:0005634 | nucleus | 2.60e-01 | 1.00e+00 | 0.396 | 7 | 158 | 3246 |
| GO:0003713 | transcription coactivator activity | 2.63e-01 | 1.00e+00 | 1.730 | 1 | 21 | 184 |
| GO:0007165 | signal transduction | 2.69e-01 | 1.00e+00 | 0.982 | 2 | 24 | 618 |
| GO:0005743 | mitochondrial inner membrane | 2.78e-01 | 1.00e+00 | 1.631 | 1 | 9 | 197 |
| GO:0005525 | GTP binding | 2.78e-01 | 1.00e+00 | 1.631 | 1 | 12 | 197 |
| GO:0006281 | DNA repair | 2.86e-01 | 1.00e+00 | 1.588 | 1 | 24 | 203 |
| GO:0008134 | transcription factor binding | 2.93e-01 | 1.00e+00 | 1.546 | 1 | 10 | 209 |
| GO:0005739 | mitochondrion | 2.94e-01 | 1.00e+00 | 0.889 | 2 | 32 | 659 |
| GO:0043234 | protein complex | 2.94e-01 | 1.00e+00 | 1.539 | 1 | 13 | 210 |
| GO:0019899 | enzyme binding | 2.94e-01 | 1.00e+00 | 1.539 | 1 | 7 | 210 |
| GO:0007186 | G-protein coupled receptor signaling pathway | 3.16e-01 | 1.00e+00 | 1.414 | 1 | 4 | 229 |
| GO:0016020 | membrane | 3.16e-01 | 1.00e+00 | 0.601 | 3 | 56 | 1207 |
| GO:0019901 | protein kinase binding | 3.20e-01 | 1.00e+00 | 1.395 | 1 | 8 | 232 |
| GO:0005730 | nucleolus | 3.21e-01 | 1.00e+00 | 0.589 | 3 | 74 | 1217 |
| GO:0007155 | cell adhesion | 3.33e-01 | 1.00e+00 | 1.323 | 1 | 8 | 244 |
| GO:0005886 | plasma membrane | 3.33e-01 | 1.00e+00 | 0.453 | 4 | 46 | 1784 |
| GO:0008270 | zinc ion binding | 3.37e-01 | 1.00e+00 | 0.748 | 2 | 39 | 727 |
| GO:0006508 | proteolysis | 3.55e-01 | 1.00e+00 | 1.209 | 1 | 11 | 264 |
| GO:0046982 | protein heterodimerization activity | 3.56e-01 | 1.00e+00 | 1.204 | 1 | 11 | 265 |
| GO:0008285 | negative regulation of cell proliferation | 3.69e-01 | 1.00e+00 | 1.140 | 1 | 11 | 277 |
| GO:0009986 | cell surface | 3.84e-01 | 1.00e+00 | 1.069 | 1 | 5 | 291 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 4.10e-01 | 1.00e+00 | 0.945 | 1 | 17 | 317 |
| GO:0043066 | negative regulation of apoptotic process | 4.15e-01 | 1.00e+00 | 0.922 | 1 | 8 | 322 |
| GO:0007596 | blood coagulation | 4.32e-01 | 1.00e+00 | 0.848 | 1 | 13 | 339 |
| GO:0005524 | ATP binding | 4.37e-01 | 1.00e+00 | 0.453 | 2 | 37 | 892 |
| GO:0048471 | perinuclear region of cytoplasm | 4.37e-01 | 1.00e+00 | 0.827 | 1 | 11 | 344 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 4.68e-01 | 1.00e+00 | 0.695 | 1 | 21 | 377 |
| GO:0003677 | DNA binding | 4.69e-01 | 1.00e+00 | 0.366 | 2 | 52 | 947 |
| GO:0045087 | innate immune response | 5.32e-01 | 1.00e+00 | 0.433 | 1 | 12 | 452 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 5.35e-01 | 1.00e+00 | 0.421 | 1 | 29 | 456 |
| GO:0005887 | integral component of plasma membrane | 6.22e-01 | 1.00e+00 | 0.086 | 1 | 8 | 575 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 6.65e-01 | 1.00e+00 | -0.078 | 1 | 29 | 644 |
| GO:0005615 | extracellular space | 6.69e-01 | 1.00e+00 | -0.093 | 1 | 15 | 651 |
| GO:0006355 | regulation of transcription, DNA-templated | 7.04e-01 | 1.00e+00 | -0.228 | 1 | 31 | 715 |
| GO:0044281 | small molecule metabolic process | 7.65e-01 | 1.00e+00 | -0.468 | 1 | 35 | 844 |