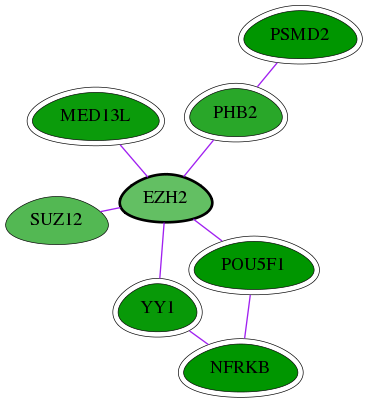
| GO:0042054 | histone methyltransferase activity | 7.53e-06 | 1.09e-01 | 8.816 | 2 | 3 | 8 |
| GO:0031011 | Ino80 complex | 2.44e-05 | 3.52e-01 | 8.009 | 2 | 6 | 14 |
| GO:0035098 | ESC/E(Z) complex | 2.82e-05 | 4.06e-01 | 7.909 | 2 | 3 | 15 |
| GO:0014013 | regulation of gliogenesis | 5.55e-04 | 1.00e+00 | 10.816 | 1 | 1 | 1 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 5.55e-04 | 1.00e+00 | 10.816 | 1 | 1 | 1 |
| GO:0045605 | negative regulation of epidermal cell differentiation | 5.55e-04 | 1.00e+00 | 10.816 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 5.55e-04 | 1.00e+00 | 10.816 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 5.55e-04 | 1.00e+00 | 10.816 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 5.55e-04 | 1.00e+00 | 10.816 | 1 | 1 | 1 |
| GO:0003723 | RNA binding | 6.77e-04 | 1.00e+00 | 3.983 | 3 | 18 | 342 |
| GO:0043565 | sequence-specific DNA binding | 7.01e-04 | 1.00e+00 | 3.966 | 3 | 11 | 346 |
| GO:0016363 | nuclear matrix | 1.08e-03 | 1.00e+00 | 5.308 | 2 | 6 | 91 |
| GO:0034696 | response to prostaglandin F | 1.11e-03 | 1.00e+00 | 9.816 | 1 | 1 | 2 |
| GO:0006403 | RNA localization | 1.11e-03 | 1.00e+00 | 9.816 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.11e-03 | 1.00e+00 | 9.816 | 1 | 1 | 2 |
| GO:0006366 | transcription from RNA polymerase II promoter | 1.22e-03 | 1.00e+00 | 3.690 | 3 | 30 | 419 |
| GO:0051154 | negative regulation of striated muscle cell differentiation | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 3 |
| GO:0070314 | G1 to G0 transition | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 3 |
| GO:0005654 | nucleoplasm | 1.73e-03 | 1.00e+00 | 2.737 | 4 | 64 | 1082 |
| GO:0046976 | histone methyltransferase activity (H3-K27 specific) | 2.22e-03 | 1.00e+00 | 8.816 | 1 | 1 | 4 |
| GO:0070734 | histone H3-K27 methylation | 2.22e-03 | 1.00e+00 | 8.816 | 1 | 1 | 4 |
| GO:0003677 | DNA binding | 2.68e-03 | 1.00e+00 | 2.566 | 4 | 49 | 1218 |
| GO:0001714 | endodermal cell fate specification | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 5 |
| GO:0071168 | protein localization to chromatin | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 5 |
| GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 5 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.96e-03 | 1.00e+00 | 3.246 | 3 | 37 | 570 |
| GO:0021695 | cerebellar cortex development | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 1 | 6 |
| GO:0060744 | mammary gland branching involved in thelarche | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 2 | 6 |
| GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 2 | 6 |
| GO:0044212 | transcription regulatory region DNA binding | 3.61e-03 | 1.00e+00 | 4.424 | 2 | 17 | 168 |
| GO:0016574 | histone ubiquitination | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 1 | 7 |
| GO:0001739 | sex chromatin | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 1 | 7 |
| GO:0045814 | negative regulation of gene expression, epigenetic | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 2 | 7 |
| GO:0005667 | transcription factor complex | 3.91e-03 | 1.00e+00 | 4.365 | 2 | 17 | 175 |
| GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 4.43e-03 | 1.00e+00 | 7.816 | 1 | 2 | 8 |
| GO:0010225 | response to UV-C | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 2 | 9 |
| GO:0000400 | four-way junction DNA binding | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 9 |
| GO:0006351 | transcription, DNA-templated | 5.06e-03 | 1.00e+00 | 2.318 | 4 | 57 | 1446 |
| GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 10 |
| GO:0035413 | positive regulation of catenin import into nucleus | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 2 | 10 |
| GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 2 | 10 |
| GO:0045120 | pronucleus | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 2 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 4 | 11 |
| GO:0005838 | proteasome regulatory particle | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 12 |
| GO:0035198 | miRNA binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 12 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 2 | 13 |
| GO:0030234 | enzyme regulator activity | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 2 | 13 |
| GO:0001824 | blastocyst development | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 15 |
| GO:0060749 | mammary gland alveolus development | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 15 |
| GO:0016571 | histone methylation | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 15 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 8.69e-03 | 1.00e+00 | 3.772 | 2 | 23 | 264 |
| GO:0042176 | regulation of protein catabolic process | 8.84e-03 | 1.00e+00 | 6.816 | 1 | 2 | 16 |
| GO:0051276 | chromosome organization | 8.84e-03 | 1.00e+00 | 6.816 | 1 | 2 | 16 |
| GO:0048593 | camera-type eye morphogenesis | 8.84e-03 | 1.00e+00 | 6.816 | 1 | 1 | 16 |
| GO:0022624 | proteasome accessory complex | 9.39e-03 | 1.00e+00 | 6.729 | 1 | 1 | 17 |
| GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 20 |
| GO:0045596 | negative regulation of cell differentiation | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 20 |
| GO:0005515 | protein binding | 1.12e-02 | 1.00e+00 | 1.067 | 7 | 198 | 6024 |
| GO:0006306 | DNA methylation | 1.21e-02 | 1.00e+00 | 6.357 | 1 | 1 | 22 |
| GO:0003682 | chromatin binding | 1.31e-02 | 1.00e+00 | 3.463 | 2 | 19 | 327 |
| GO:0010718 | positive regulation of epithelial to mesenchymal transition | 1.38e-02 | 1.00e+00 | 6.172 | 1 | 4 | 25 |
| GO:0031519 | PcG protein complex | 1.38e-02 | 1.00e+00 | 6.172 | 1 | 1 | 25 |
| GO:0006355 | regulation of transcription, DNA-templated | 1.42e-02 | 1.00e+00 | 2.440 | 3 | 43 | 997 |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1.43e-02 | 1.00e+00 | 6.116 | 1 | 2 | 26 |
| GO:0030331 | estrogen receptor binding | 1.49e-02 | 1.00e+00 | 6.061 | 1 | 2 | 27 |
| GO:0005634 | nucleus | 1.49e-02 | 1.00e+00 | 1.247 | 6 | 159 | 4559 |
| GO:0034644 | cellular response to UV | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 30 |
| GO:0032320 | positive regulation of Ras GTPase activity | 1.76e-02 | 1.00e+00 | 5.816 | 1 | 1 | 32 |
| GO:0001104 | RNA polymerase II transcription cofactor activity | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 8 | 33 |
| GO:0016592 | mediator complex | 1.93e-02 | 1.00e+00 | 5.687 | 1 | 10 | 35 |
| GO:0042752 | regulation of circadian rhythm | 2.03e-02 | 1.00e+00 | 5.607 | 1 | 3 | 37 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.04e-02 | 1.00e+00 | 3.126 | 2 | 24 | 413 |
| GO:0031490 | chromatin DNA binding | 2.09e-02 | 1.00e+00 | 5.568 | 1 | 2 | 38 |
| GO:0035019 | somatic stem cell maintenance | 2.20e-02 | 1.00e+00 | 5.494 | 1 | 6 | 40 |
| GO:0070301 | cellular response to hydrogen peroxide | 2.25e-02 | 1.00e+00 | 5.459 | 1 | 2 | 41 |
| GO:0001047 | core promoter binding | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 5 | 45 |
| GO:0048511 | rhythmic process | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 4 | 45 |
| GO:0035064 | methylated histone binding | 2.52e-02 | 1.00e+00 | 5.293 | 1 | 2 | 46 |
| GO:0043406 | positive regulation of MAP kinase activity | 2.52e-02 | 1.00e+00 | 5.293 | 1 | 2 | 46 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 2.74e-02 | 1.00e+00 | 5.172 | 1 | 1 | 50 |
| GO:0009611 | response to wounding | 2.85e-02 | 1.00e+00 | 5.116 | 1 | 5 | 52 |
| GO:0000724 | double-strand break repair via homologous recombination | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 2 | 55 |
| GO:0000502 | proteasome complex | 3.17e-02 | 1.00e+00 | 4.958 | 1 | 3 | 58 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 3.23e-02 | 1.00e+00 | 4.933 | 1 | 5 | 59 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 3.50e-02 | 1.00e+00 | 4.816 | 1 | 1 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 2 | 65 |
| GO:0006310 | DNA recombination | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 7 | 65 |
| GO:0010468 | regulation of gene expression | 3.71e-02 | 1.00e+00 | 4.729 | 1 | 2 | 68 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 3.82e-02 | 1.00e+00 | 4.687 | 1 | 2 | 70 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 3.93e-02 | 1.00e+00 | 4.646 | 1 | 1 | 72 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 4.03e-02 | 1.00e+00 | 4.607 | 1 | 4 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.03e-02 | 1.00e+00 | 4.607 | 1 | 2 | 74 |
| GO:0002020 | protease binding | 4.03e-02 | 1.00e+00 | 4.607 | 1 | 6 | 74 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 4.09e-02 | 1.00e+00 | 4.587 | 1 | 1 | 75 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 4.30e-02 | 1.00e+00 | 4.512 | 1 | 2 | 79 |
| GO:0009653 | anatomical structure morphogenesis | 4.51e-02 | 1.00e+00 | 4.441 | 1 | 2 | 83 |
| GO:0009952 | anterior/posterior pattern specification | 4.51e-02 | 1.00e+00 | 4.441 | 1 | 4 | 83 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 4.99e-02 | 1.00e+00 | 4.293 | 1 | 1 | 92 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 5.41e-02 | 1.00e+00 | 2.367 | 2 | 39 | 699 |
| GO:0042127 | regulation of cell proliferation | 5.89e-02 | 1.00e+00 | 4.048 | 1 | 4 | 109 |
| GO:0000209 | protein polyubiquitination | 6.26e-02 | 1.00e+00 | 3.958 | 1 | 3 | 116 |
| GO:0006325 | chromatin organization | 6.36e-02 | 1.00e+00 | 3.933 | 1 | 12 | 118 |
| GO:0000790 | nuclear chromatin | 6.94e-02 | 1.00e+00 | 3.805 | 1 | 6 | 129 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 7.97e-02 | 1.00e+00 | 3.597 | 1 | 3 | 149 |
| GO:0042981 | regulation of apoptotic process | 8.03e-02 | 1.00e+00 | 3.587 | 1 | 4 | 150 |
| GO:0006974 | cellular response to DNA damage stimulus | 8.08e-02 | 1.00e+00 | 3.578 | 1 | 7 | 151 |
| GO:0034641 | cellular nitrogen compound metabolic process | 9.10e-02 | 1.00e+00 | 3.398 | 1 | 2 | 171 |
| GO:0031625 | ubiquitin protein ligase binding | 9.46e-02 | 1.00e+00 | 3.340 | 1 | 5 | 178 |
| GO:0003714 | transcription corepressor activity | 9.46e-02 | 1.00e+00 | 3.340 | 1 | 11 | 178 |
| GO:0016071 | mRNA metabolic process | 1.17e-01 | 1.00e+00 | 3.015 | 1 | 8 | 223 |
| GO:0003713 | transcription coactivator activity | 1.24e-01 | 1.00e+00 | 2.927 | 1 | 24 | 237 |
| GO:0008134 | transcription factor binding | 1.27e-01 | 1.00e+00 | 2.897 | 1 | 18 | 242 |
| GO:0016070 | RNA metabolic process | 1.29e-01 | 1.00e+00 | 2.868 | 1 | 8 | 247 |
| GO:0007283 | spermatogenesis | 1.36e-01 | 1.00e+00 | 2.783 | 1 | 3 | 262 |
| GO:0006281 | DNA repair | 1.36e-01 | 1.00e+00 | 2.788 | 1 | 14 | 261 |
| GO:0005743 | mitochondrial inner membrane | 1.44e-01 | 1.00e+00 | 2.702 | 1 | 6 | 277 |
| GO:0006954 | inflammatory response | 1.44e-01 | 1.00e+00 | 2.702 | 1 | 6 | 277 |
| GO:0043234 | protein complex | 1.52e-01 | 1.00e+00 | 2.612 | 1 | 11 | 295 |
| GO:0030154 | cell differentiation | 1.55e-01 | 1.00e+00 | 2.582 | 1 | 5 | 301 |
| GO:0008284 | positive regulation of cell proliferation | 1.93e-01 | 1.00e+00 | 2.239 | 1 | 10 | 382 |
| GO:0000278 | mitotic cell cycle | 1.97e-01 | 1.00e+00 | 2.205 | 1 | 15 | 391 |
| GO:0043066 | negative regulation of apoptotic process | 2.12e-01 | 1.00e+00 | 2.088 | 1 | 16 | 424 |
| GO:0005730 | nucleolus | 2.29e-01 | 1.00e+00 | 1.136 | 2 | 66 | 1641 |
| GO:0016032 | viral process | 2.61e-01 | 1.00e+00 | 1.755 | 1 | 26 | 534 |
| GO:0006915 | apoptotic process | 2.69e-01 | 1.00e+00 | 1.700 | 1 | 12 | 555 |
| GO:0010467 | gene expression | 3.16e-01 | 1.00e+00 | 1.430 | 1 | 45 | 669 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 3.62e-01 | 1.00e+00 | 1.192 | 1 | 41 | 789 |
| GO:0070062 | extracellular vesicular exosome | 3.95e-01 | 1.00e+00 | 0.587 | 2 | 57 | 2400 |
| GO:0005829 | cytosol | 4.15e-01 | 1.00e+00 | 0.531 | 2 | 86 | 2496 |
| GO:0008270 | zinc ion binding | 4.36e-01 | 1.00e+00 | 0.855 | 1 | 27 | 997 |
| GO:0005739 | mitochondrion | 4.37e-01 | 1.00e+00 | 0.853 | 1 | 23 | 998 |
| GO:0044822 | poly(A) RNA binding | 4.56e-01 | 1.00e+00 | 0.772 | 1 | 50 | 1056 |
| GO:0044281 | small molecule metabolic process | 5.04e-01 | 1.00e+00 | 0.574 | 1 | 20 | 1211 |
| GO:0046872 | metal ion binding | 5.32e-01 | 1.00e+00 | 0.464 | 1 | 29 | 1307 |
| GO:0016020 | membrane | 6.29e-01 | 1.00e+00 | 0.101 | 1 | 46 | 1681 |
| GO:0005737 | cytoplasm | 6.60e-01 | 1.00e+00 | -0.063 | 2 | 124 | 3767 |