| GO:0001933 | negative regulation of protein phosphorylation | 6.97e-04 | 1.00e+00 | 5.646 | 2 | 3 | 30 |
| GO:0015036 | disulfide oxidoreductase activity | 1.33e-03 | 1.00e+00 | 9.553 | 1 | 1 | 1 |
| GO:0001776 | leukocyte homeostasis | 1.33e-03 | 1.00e+00 | 9.553 | 1 | 1 | 1 |
| GO:0034975 | protein folding in endoplasmic reticulum | 1.33e-03 | 1.00e+00 | 9.553 | 1 | 1 | 1 |
| GO:0006281 | DNA repair | 2.17e-03 | 1.00e+00 | 3.473 | 3 | 24 | 203 |
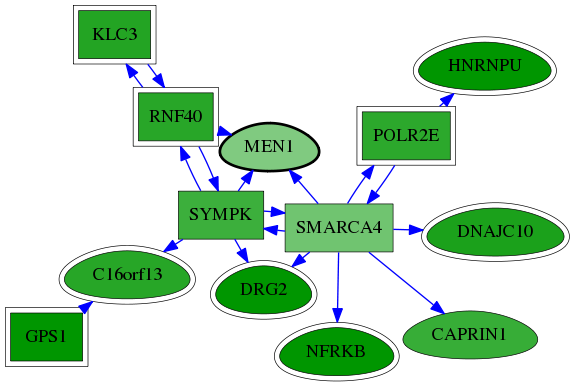
| GO:0006338 | chromatin remodeling | 2.33e-03 | 1.00e+00 | 4.772 | 2 | 7 | 55 |
| GO:0001832 | blastocyst growth | 2.66e-03 | 1.00e+00 | 8.553 | 1 | 1 | 2 |
| GO:0007403 | glial cell fate determination | 2.66e-03 | 1.00e+00 | 8.553 | 1 | 1 | 2 |
| GO:0034663 | endoplasmic reticulum chaperone complex | 2.66e-03 | 1.00e+00 | 8.553 | 1 | 1 | 2 |
| GO:0035887 | aortic smooth muscle cell differentiation | 2.66e-03 | 1.00e+00 | 8.553 | 1 | 2 | 2 |
| GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | 2.66e-03 | 1.00e+00 | 8.553 | 1 | 1 | 2 |
| GO:0007070 | negative regulation of transcription from RNA polymerase II promoter during mitosis | 2.66e-03 | 1.00e+00 | 8.553 | 1 | 1 | 2 |
| GO:0016020 | membrane | 2.79e-03 | 1.00e+00 | 1.901 | 6 | 56 | 1207 |
| GO:0047485 | protein N-terminus binding | 3.25e-03 | 1.00e+00 | 4.531 | 2 | 6 | 65 |
| GO:0031062 | positive regulation of histone methylation | 3.99e-03 | 1.00e+00 | 7.968 | 1 | 1 | 3 |
| GO:0001835 | blastocyst hatching | 3.99e-03 | 1.00e+00 | 7.968 | 1 | 1 | 3 |
| GO:0010424 | DNA methylation on cytosine within a CG sequence | 3.99e-03 | 1.00e+00 | 7.968 | 1 | 1 | 3 |
| GO:0005726 | perichromatin fibrils | 3.99e-03 | 1.00e+00 | 7.968 | 1 | 1 | 3 |
| GO:0003407 | neural retina development | 5.32e-03 | 1.00e+00 | 7.553 | 1 | 2 | 4 |
| GO:0048562 | embryonic organ morphogenesis | 5.32e-03 | 1.00e+00 | 7.553 | 1 | 1 | 4 |
| GO:0060318 | definitive erythrocyte differentiation | 5.32e-03 | 1.00e+00 | 7.553 | 1 | 1 | 4 |
| GO:0032925 | regulation of activin receptor signaling pathway | 5.32e-03 | 1.00e+00 | 7.553 | 1 | 1 | 4 |
| GO:0033503 | HULC complex | 5.32e-03 | 1.00e+00 | 7.553 | 1 | 1 | 4 |
| GO:0002051 | osteoblast fate commitment | 5.32e-03 | 1.00e+00 | 7.553 | 1 | 1 | 4 |
| GO:0051974 | negative regulation of telomerase activity | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 5 |
| GO:0070307 | lens fiber cell development | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 2 | 5 |
| GO:0030957 | Tat protein binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 5 |
| GO:0000403 | Y-form DNA binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 5 |
| GO:0048730 | epidermis morphogenesis | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 5 |
| GO:0061003 | positive regulation of dendritic spine morphogenesis | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 5 |
| GO:0001055 | RNA polymerase II activity | 7.96e-03 | 1.00e+00 | 6.968 | 1 | 3 | 6 |
| GO:0006346 | methylation-dependent chromatin silencing | 7.96e-03 | 1.00e+00 | 6.968 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 7.96e-03 | 1.00e+00 | 6.968 | 1 | 2 | 6 |
| GO:0001671 | ATPase activator activity | 7.96e-03 | 1.00e+00 | 6.968 | 1 | 1 | 6 |
| GO:0000400 | four-way junction DNA binding | 9.28e-03 | 1.00e+00 | 6.746 | 1 | 3 | 7 |
| GO:0033523 | histone H2B ubiquitination | 9.28e-03 | 1.00e+00 | 6.746 | 1 | 1 | 7 |
| GO:0043005 | neuron projection | 9.53e-03 | 1.00e+00 | 3.733 | 2 | 6 | 113 |
| GO:0051787 | misfolded protein binding | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 2 | 8 |
| GO:0031011 | Ino80 complex | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 3 | 8 |
| GO:0008088 | axon cargo transport | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 1 | 8 |
| GO:0010388 | cullin deneddylation | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 4 | 8 |
| GO:0070182 | DNA polymerase binding | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 1 | 8 |
| GO:0010390 | histone monoubiquitination | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 2 | 8 |
| GO:0046621 | negative regulation of organ growth | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 1 | 9 |
| GO:0034968 | histone lysine methylation | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 1 | 9 |
| GO:0035253 | ciliary rootlet | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 1 | 9 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.21e-02 | 1.00e+00 | 3.553 | 2 | 15 | 128 |
| GO:0043923 | positive regulation by host of viral transcription | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 10 |
| GO:0005736 | DNA-directed RNA polymerase I complex | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 4 | 10 |
| GO:0005095 | GTPase inhibitor activity | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 10 |
| GO:0071564 | npBAF complex | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 3 | 10 |
| GO:0035097 | histone methyltransferase complex | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 10 |
| GO:0001054 | RNA polymerase I activity | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 4 | 10 |
| GO:0035307 | positive regulation of protein dephosphorylation | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 10 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 1.46e-02 | 1.00e+00 | 6.094 | 1 | 4 | 11 |
| GO:0071565 | nBAF complex | 1.46e-02 | 1.00e+00 | 6.094 | 1 | 3 | 11 |
| GO:0050775 | positive regulation of dendrite morphogenesis | 1.46e-02 | 1.00e+00 | 6.094 | 1 | 1 | 11 |
| GO:0030544 | Hsp70 protein binding | 1.59e-02 | 1.00e+00 | 5.968 | 1 | 1 | 12 |
| GO:0070577 | lysine-acetylated histone binding | 1.59e-02 | 1.00e+00 | 5.968 | 1 | 2 | 12 |
| GO:0017075 | syntaxin-1 binding | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 1 | 13 |
| GO:0006337 | nucleosome disassembly | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 2 | 13 |
| GO:0016514 | SWI/SNF complex | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 3 | 13 |
| GO:0005666 | DNA-directed RNA polymerase III complex | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 3 | 13 |
| GO:0005665 | DNA-directed RNA polymerase II, core complex | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 3 | 13 |
| GO:0001056 | RNA polymerase III activity | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 3 | 13 |
| GO:0015035 | protein disulfide oxidoreductase activity | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 1 | 13 |
| GO:0060347 | heart trabecula formation | 1.72e-02 | 1.00e+00 | 5.853 | 1 | 1 | 13 |
| GO:0006386 | termination of RNA polymerase III transcription | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 14 |
| GO:0060135 | maternal process involved in female pregnancy | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 14 |
| GO:0030902 | hindbrain development | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 2 | 14 |
| GO:0006385 | transcription elongation from RNA polymerase III promoter | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 14 |
| GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 3 | 14 |
| GO:0031514 | motile cilium | 1.85e-02 | 1.00e+00 | 5.746 | 1 | 1 | 14 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.87e-02 | 1.00e+00 | 3.222 | 2 | 18 | 161 |
| GO:0008380 | RNA splicing | 1.96e-02 | 1.00e+00 | 3.187 | 2 | 21 | 165 |
| GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 1.98e-02 | 1.00e+00 | 5.646 | 1 | 2 | 15 |
| GO:0032781 | positive regulation of ATPase activity | 1.98e-02 | 1.00e+00 | 5.646 | 1 | 2 | 15 |
| GO:0002076 | osteoblast development | 1.98e-02 | 1.00e+00 | 5.646 | 1 | 1 | 15 |
| GO:0000188 | inactivation of MAPK activity | 1.98e-02 | 1.00e+00 | 5.646 | 1 | 1 | 15 |
| GO:0018024 | histone-lysine N-methyltransferase activity | 1.98e-02 | 1.00e+00 | 5.646 | 1 | 1 | 15 |
| GO:0005719 | nuclear euchromatin | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 2 | 16 |
| GO:0000792 | heterochromatin | 2.24e-02 | 1.00e+00 | 5.466 | 1 | 1 | 17 |
| GO:0003899 | DNA-directed RNA polymerase activity | 2.37e-02 | 1.00e+00 | 5.383 | 1 | 3 | 18 |
| GO:0070412 | R-SMAD binding | 2.37e-02 | 1.00e+00 | 5.383 | 1 | 1 | 18 |
| GO:0006378 | mRNA polyadenylation | 2.37e-02 | 1.00e+00 | 5.383 | 1 | 3 | 18 |
| GO:0035116 | embryonic hindlimb morphogenesis | 2.37e-02 | 1.00e+00 | 5.383 | 1 | 2 | 18 |
| GO:0005654 | nucleoplasm | 2.38e-02 | 1.00e+00 | 1.778 | 4 | 68 | 876 |
| GO:0046329 | negative regulation of JNK cascade | 2.50e-02 | 1.00e+00 | 5.305 | 1 | 1 | 19 |
| GO:0001105 | RNA polymerase II transcription coactivator activity | 2.50e-02 | 1.00e+00 | 5.305 | 1 | 2 | 19 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 2.50e-02 | 1.00e+00 | 5.305 | 1 | 1 | 19 |
| GO:0043044 | ATP-dependent chromatin remodeling | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 3 | 20 |
| GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 1 | 20 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 1 | 20 |
| GO:0043388 | positive regulation of DNA binding | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 2 | 21 |
| GO:0019894 | kinesin binding | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 1 | 21 |
| GO:0051781 | positive regulation of cell division | 2.89e-02 | 1.00e+00 | 5.094 | 1 | 2 | 22 |
| GO:0045668 | negative regulation of osteoblast differentiation | 2.89e-02 | 1.00e+00 | 5.094 | 1 | 1 | 22 |
| GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process | 2.89e-02 | 1.00e+00 | 5.094 | 1 | 1 | 22 |
| GO:0030177 | positive regulation of Wnt signaling pathway | 3.02e-02 | 1.00e+00 | 5.029 | 1 | 1 | 23 |
| GO:0043234 | protein complex | 3.07e-02 | 1.00e+00 | 2.839 | 2 | 13 | 210 |
| GO:0010467 | gene expression | 3.10e-02 | 1.00e+00 | 2.075 | 3 | 49 | 535 |
| GO:0006370 | 7-methylguanosine mRNA capping | 3.15e-02 | 1.00e+00 | 4.968 | 1 | 5 | 24 |
| GO:0008180 | COP9 signalosome | 3.15e-02 | 1.00e+00 | 4.968 | 1 | 6 | 24 |
| GO:0051117 | ATPase binding | 3.15e-02 | 1.00e+00 | 4.968 | 1 | 1 | 24 |
| GO:0031492 | nucleosomal DNA binding | 3.28e-02 | 1.00e+00 | 4.909 | 1 | 3 | 25 |
| GO:0005634 | nucleus | 3.40e-02 | 1.00e+00 | 0.889 | 8 | 158 | 3246 |
| GO:0010332 | response to gamma radiation | 3.41e-02 | 1.00e+00 | 4.853 | 1 | 2 | 26 |
| GO:0010494 | cytoplasmic stress granule | 3.41e-02 | 1.00e+00 | 4.853 | 1 | 4 | 26 |
| GO:0006360 | transcription from RNA polymerase I promoter | 3.54e-02 | 1.00e+00 | 4.798 | 1 | 4 | 27 |
| GO:0043966 | histone H3 acetylation | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 4 | 28 |
| GO:0048704 | embryonic skeletal system morphogenesis | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 1 | 28 |
| GO:0008094 | DNA-dependent ATPase activity | 3.79e-02 | 1.00e+00 | 4.695 | 1 | 6 | 29 |
| GO:0006383 | transcription from RNA polymerase III promoter | 3.79e-02 | 1.00e+00 | 4.695 | 1 | 6 | 29 |
| GO:0045786 | negative regulation of cell cycle | 3.92e-02 | 1.00e+00 | 4.646 | 1 | 3 | 30 |
| GO:0050681 | androgen receptor binding | 3.92e-02 | 1.00e+00 | 4.646 | 1 | 2 | 30 |
| GO:0032154 | cleavage furrow | 3.92e-02 | 1.00e+00 | 4.646 | 1 | 3 | 30 |
| GO:0034976 | response to endoplasmic reticulum stress | 4.05e-02 | 1.00e+00 | 4.599 | 1 | 2 | 31 |
| GO:0045454 | cell redox homeostasis | 4.05e-02 | 1.00e+00 | 4.599 | 1 | 3 | 31 |
| GO:0005871 | kinesin complex | 4.05e-02 | 1.00e+00 | 4.599 | 1 | 3 | 31 |
| GO:0003723 | RNA binding | 4.14e-02 | 1.00e+00 | 2.605 | 2 | 22 | 247 |
| GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 4.18e-02 | 1.00e+00 | 4.553 | 1 | 1 | 32 |
| GO:0017148 | negative regulation of translation | 4.31e-02 | 1.00e+00 | 4.509 | 1 | 5 | 33 |
| GO:0032092 | positive regulation of protein binding | 4.44e-02 | 1.00e+00 | 4.466 | 1 | 2 | 34 |
| GO:0007254 | JNK cascade | 4.44e-02 | 1.00e+00 | 4.466 | 1 | 1 | 34 |
| GO:0000902 | cell morphogenesis | 4.56e-02 | 1.00e+00 | 4.424 | 1 | 5 | 35 |
| GO:0050434 | positive regulation of viral transcription | 4.56e-02 | 1.00e+00 | 4.424 | 1 | 5 | 35 |
| GO:0005515 | protein binding | 4.61e-02 | 1.00e+00 | 0.713 | 9 | 198 | 4124 |
| GO:0030900 | forebrain development | 4.69e-02 | 1.00e+00 | 4.383 | 1 | 2 | 36 |
| GO:0009411 | response to UV | 4.69e-02 | 1.00e+00 | 4.383 | 1 | 3 | 36 |
| GO:0030674 | protein binding, bridging | 4.82e-02 | 1.00e+00 | 4.344 | 1 | 1 | 37 |
| GO:0030216 | keratinocyte differentiation | 4.95e-02 | 1.00e+00 | 4.305 | 1 | 1 | 38 |
| GO:0019827 | stem cell maintenance | 5.07e-02 | 1.00e+00 | 4.268 | 1 | 4 | 39 |
| GO:0003777 | microtubule motor activity | 5.07e-02 | 1.00e+00 | 4.268 | 1 | 3 | 39 |
| GO:0006396 | RNA processing | 5.20e-02 | 1.00e+00 | 4.231 | 1 | 4 | 40 |
| GO:0006283 | transcription-coupled nucleotide-excision repair | 5.20e-02 | 1.00e+00 | 4.231 | 1 | 6 | 40 |
| GO:0045669 | positive regulation of osteoblast differentiation | 5.33e-02 | 1.00e+00 | 4.195 | 1 | 2 | 41 |
| GO:0004386 | helicase activity | 5.45e-02 | 1.00e+00 | 4.161 | 1 | 3 | 42 |
| GO:0000932 | cytoplasmic mRNA processing body | 5.58e-02 | 1.00e+00 | 4.127 | 1 | 3 | 43 |
| GO:0051087 | chaperone binding | 5.83e-02 | 1.00e+00 | 4.061 | 1 | 3 | 45 |
| GO:0001570 | vasculogenesis | 5.83e-02 | 1.00e+00 | 4.061 | 1 | 2 | 45 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 5.83e-02 | 1.00e+00 | 4.061 | 1 | 2 | 45 |
| GO:0002039 | p53 binding | 5.83e-02 | 1.00e+00 | 4.061 | 1 | 4 | 45 |
| GO:0060021 | palate development | 6.08e-02 | 1.00e+00 | 3.998 | 1 | 1 | 47 |
| GO:0032481 | positive regulation of type I interferon production | 6.21e-02 | 1.00e+00 | 3.968 | 1 | 3 | 48 |
| GO:0000151 | ubiquitin ligase complex | 6.21e-02 | 1.00e+00 | 3.968 | 1 | 4 | 48 |
| GO:0030097 | hemopoiesis | 6.21e-02 | 1.00e+00 | 3.968 | 1 | 3 | 48 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 6.33e-02 | 1.00e+00 | 3.938 | 1 | 4 | 49 |
| GO:0006310 | DNA recombination | 6.46e-02 | 1.00e+00 | 3.909 | 1 | 8 | 50 |
| GO:0002020 | protease binding | 6.46e-02 | 1.00e+00 | 3.909 | 1 | 3 | 50 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 6.47e-02 | 1.00e+00 | 2.245 | 2 | 17 | 317 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 6.71e-02 | 1.00e+00 | 3.853 | 1 | 8 | 52 |
| GO:0005730 | nucleolus | 6.82e-02 | 1.00e+00 | 1.304 | 4 | 74 | 1217 |
| GO:0006289 | nucleotide-excision repair | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 6 | 53 |
| GO:0006366 | transcription from RNA polymerase II promoter | 7.36e-02 | 1.00e+00 | 2.139 | 2 | 23 | 341 |
| GO:0000165 | MAPK cascade | 7.46e-02 | 1.00e+00 | 3.695 | 1 | 3 | 58 |
| GO:0000785 | chromatin | 7.58e-02 | 1.00e+00 | 3.670 | 1 | 6 | 59 |
| GO:0071013 | catalytic step 2 spliceosome | 7.83e-02 | 1.00e+00 | 3.622 | 1 | 5 | 61 |
| GO:0005923 | tight junction | 8.20e-02 | 1.00e+00 | 3.553 | 1 | 2 | 64 |
| GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity | 8.57e-02 | 1.00e+00 | 3.487 | 1 | 5 | 67 |
| GO:0030308 | negative regulation of cell growth | 8.93e-02 | 1.00e+00 | 3.424 | 1 | 3 | 70 |
| GO:0001889 | liver development | 9.06e-02 | 1.00e+00 | 3.403 | 1 | 4 | 71 |
| GO:0016363 | nuclear matrix | 9.30e-02 | 1.00e+00 | 3.363 | 1 | 10 | 73 |
| GO:0001649 | osteoblast differentiation | 9.42e-02 | 1.00e+00 | 3.344 | 1 | 5 | 74 |
| GO:0030529 | ribonucleoprotein complex | 1.04e-01 | 1.00e+00 | 3.195 | 1 | 5 | 82 |
| GO:0003690 | double-stranded DNA binding | 1.05e-01 | 1.00e+00 | 3.178 | 1 | 6 | 83 |
| GO:0005737 | cytoplasm | 1.09e-01 | 1.00e+00 | 0.775 | 6 | 127 | 2633 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 1.16e-01 | 1.00e+00 | 3.029 | 1 | 6 | 92 |
| GO:0007050 | cell cycle arrest | 1.16e-01 | 1.00e+00 | 3.029 | 1 | 4 | 92 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 1.21e-01 | 1.00e+00 | 1.720 | 2 | 29 | 456 |
| GO:0008017 | microtubule binding | 1.22e-01 | 1.00e+00 | 2.953 | 1 | 9 | 97 |
| GO:0003677 | DNA binding | 1.25e-01 | 1.00e+00 | 1.251 | 3 | 52 | 947 |
| GO:0005788 | endoplasmic reticulum lumen | 1.30e-01 | 1.00e+00 | 2.853 | 1 | 4 | 104 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 1.34e-01 | 1.00e+00 | 2.811 | 1 | 5 | 107 |
| GO:0000790 | nuclear chromatin | 1.36e-01 | 1.00e+00 | 2.785 | 1 | 9 | 109 |
| GO:0007049 | cell cycle | 1.43e-01 | 1.00e+00 | 2.707 | 1 | 8 | 115 |
| GO:0007420 | brain development | 1.47e-01 | 1.00e+00 | 2.658 | 1 | 3 | 119 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.50e-01 | 1.00e+00 | 2.634 | 1 | 11 | 121 |
| GO:0032403 | protein complex binding | 1.60e-01 | 1.00e+00 | 2.531 | 1 | 5 | 130 |
| GO:0003714 | transcription corepressor activity | 1.68e-01 | 1.00e+00 | 2.455 | 1 | 5 | 137 |
| GO:0031625 | ubiquitin protein ligase binding | 1.69e-01 | 1.00e+00 | 2.444 | 1 | 7 | 138 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.73e-01 | 1.00e+00 | 2.403 | 1 | 7 | 142 |
| GO:0044212 | transcription regulatory region DNA binding | 1.75e-01 | 1.00e+00 | 2.393 | 1 | 13 | 143 |
| GO:0016874 | ligase activity | 1.78e-01 | 1.00e+00 | 2.363 | 1 | 11 | 146 |
| GO:0005874 | microtubule | 1.97e-01 | 1.00e+00 | 2.204 | 1 | 10 | 163 |
| GO:0030425 | dendrite | 2.00e-01 | 1.00e+00 | 2.178 | 1 | 2 | 166 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 2.10e-01 | 1.00e+00 | 1.222 | 2 | 29 | 644 |
| GO:0004842 | ubiquitin-protein transferase activity | 2.14e-01 | 1.00e+00 | 2.069 | 1 | 17 | 179 |
| GO:0008152 | metabolic process | 2.17e-01 | 1.00e+00 | 2.045 | 1 | 6 | 182 |
| GO:0003713 | transcription coactivator activity | 2.19e-01 | 1.00e+00 | 2.029 | 1 | 21 | 184 |
| GO:0006954 | inflammatory response | 2.20e-01 | 1.00e+00 | 2.022 | 1 | 3 | 185 |
| GO:0005856 | cytoskeleton | 2.26e-01 | 1.00e+00 | 1.983 | 1 | 8 | 190 |
| GO:0005525 | GTP binding | 2.33e-01 | 1.00e+00 | 1.931 | 1 | 12 | 197 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 2.39e-01 | 1.00e+00 | 1.888 | 1 | 14 | 203 |
| GO:0006200 | ATP catabolic process | 2.56e-01 | 1.00e+00 | 1.778 | 1 | 13 | 219 |
| GO:0030198 | extracellular matrix organization | 2.58e-01 | 1.00e+00 | 1.765 | 1 | 7 | 221 |
| GO:0007155 | cell adhesion | 2.80e-01 | 1.00e+00 | 1.622 | 1 | 8 | 244 |
| GO:0003682 | chromatin binding | 2.88e-01 | 1.00e+00 | 1.576 | 1 | 15 | 252 |
| GO:0044822 | poly(A) RNA binding | 2.88e-01 | 1.00e+00 | 0.911 | 2 | 45 | 799 |
| GO:0043565 | sequence-specific DNA binding | 3.10e-01 | 1.00e+00 | 1.450 | 1 | 8 | 275 |
| GO:0008285 | negative regulation of cell proliferation | 3.12e-01 | 1.00e+00 | 1.439 | 1 | 11 | 277 |
| GO:0009986 | cell surface | 3.25e-01 | 1.00e+00 | 1.368 | 1 | 5 | 291 |
| GO:0005524 | ATP binding | 3.36e-01 | 1.00e+00 | 0.752 | 2 | 37 | 892 |
| GO:0055114 | oxidation-reduction process | 3.41e-01 | 1.00e+00 | 1.286 | 1 | 9 | 308 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 4.01e-01 | 1.00e+00 | 0.995 | 1 | 21 | 377 |
| GO:0005783 | endoplasmic reticulum | 4.04e-01 | 1.00e+00 | 0.979 | 1 | 12 | 381 |
| GO:0006351 | transcription, DNA-templated | 4.28e-01 | 1.00e+00 | 0.482 | 2 | 47 | 1076 |
| GO:0005829 | cytosol | 4.35e-01 | 1.00e+00 | 0.335 | 3 | 88 | 1787 |
| GO:0042803 | protein homodimerization activity | 4.42e-01 | 1.00e+00 | 0.811 | 1 | 13 | 428 |
| GO:0016032 | viral process | 4.42e-01 | 1.00e+00 | 0.811 | 1 | 30 | 428 |
| GO:0045087 | innate immune response | 4.60e-01 | 1.00e+00 | 0.733 | 1 | 12 | 452 |
| GO:0005887 | integral component of plasma membrane | 5.46e-01 | 1.00e+00 | 0.386 | 1 | 8 | 575 |
| GO:0007165 | signal transduction | 5.73e-01 | 1.00e+00 | 0.282 | 1 | 24 | 618 |
| GO:0005615 | extracellular space | 5.92e-01 | 1.00e+00 | 0.206 | 1 | 15 | 651 |
| GO:0005739 | mitochondrion | 5.97e-01 | 1.00e+00 | 0.189 | 1 | 32 | 659 |
| GO:0006355 | regulation of transcription, DNA-templated | 6.28e-01 | 1.00e+00 | 0.071 | 1 | 31 | 715 |
| GO:0008270 | zinc ion binding | 6.34e-01 | 1.00e+00 | 0.047 | 1 | 39 | 727 |
| GO:0005886 | plasma membrane | 9.28e-01 | 1.00e+00 | -1.248 | 1 | 46 | 1784 |