int-snw-55161
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.994 | 2.86e-17 | 6.27e-05 | 7.97e-04 |
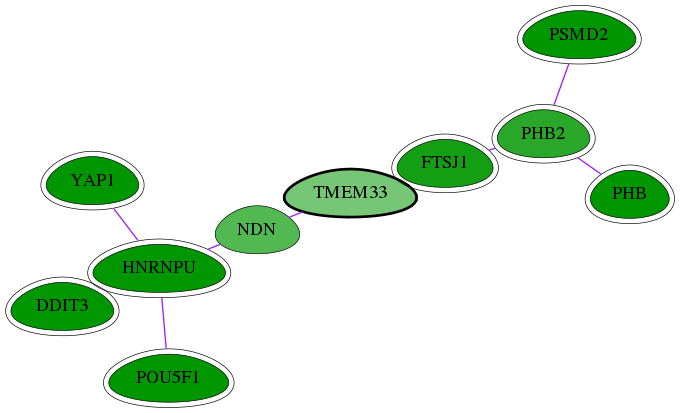
Genes (10)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| PSMD2 | 5708 | 47 | -4.172 | 3.157 | 386 | Yes | Yes |
| PHB2 | 11331 | 5 | -2.236 | 2.994 | 140 | Yes | - |
| PHB | 5245 | 15 | -2.743 | 3.004 | 127 | Yes | Yes |
| NDN | 4692 | 1 | -1.801 | 2.994 | 32 | - | - |
| FTSJ1 | 24140 | 4 | -2.463 | 2.994 | 95 | Yes | Yes |
| YAP1 | 10413 | 94 | -4.256 | 3.538 | 53 | Yes | - |
| HNRNPU | 3192 | 76 | -2.869 | 3.538 | 139 | Yes | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| DDIT3 | 1649 | 60 | -2.825 | 3.504 | 343 | Yes | Yes |
| [ TMEM33 ] | 55161 | 1 | -1.429 | 2.994 | 26 | - | - |
Interactions (9)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| PHB2 | 11331 | FTSJ1 | 24140 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| HNRNPU | 3192 | YAP1 | 10413 | pp | -- | int.I2D: HPRD; int.HPRD: in vitro, in vivo |
| HNRNPU | 3192 | NDN | 4692 | pp | -- | int.I2D: BioGrid, HPRD; int.HPRD: in vitro |
| PHB | 5245 | PHB2 | 11331 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast, IntAct_Yeast |
| HNRNPU | 3192 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| DDIT3 | 1649 | HNRNPU | 3192 | pp | -- | int.I2D: BEHRENDS_AUTOPHAGY_LOW |
| NDN | 4692 | TMEM33 | 55161 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: IntAct, BioGrid |
| FTSJ1 | 24140 | TMEM33 | 55161 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| PSMD2 | 5708 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
Related GO terms (158)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0044212 | transcription regulatory region DNA binding | 3.53e-06 | 5.09e-02 | 5.102 | 4 | 17 | 168 |
| GO:0005654 | nucleoplasm | 2.84e-05 | 4.09e-01 | 3.000 | 6 | 64 | 1082 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 3.36e-05 | 4.85e-01 | 7.794 | 2 | 2 | 13 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.94e-04 | 1.00e+00 | 2.855 | 5 | 43 | 997 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0002128 | tRNA nucleoside ribose methylation | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:2000016 | negative regulation of determination of dorsal identity | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0060242 | contact inhibition | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0008175 | tRNA methyltransferase activity | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0044324 | regulation of transcription involved in anterior/posterior axis specification | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0001649 | osteoblast differentiation | 1.75e-03 | 1.00e+00 | 4.971 | 2 | 5 | 92 |
| GO:0048871 | multicellular organismal homeostasis | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.41e-03 | 1.00e+00 | 3.389 | 3 | 24 | 413 |
| GO:0001714 | endodermal cell fate specification | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0043620 | regulation of DNA-templated transcription in response to stress | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 2 | 5 |
| GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0050847 | progesterone receptor signaling pathway | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 2 | 6 |
| GO:0002181 | cytoplasmic translation | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0071514 | genetic imprinting | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0060744 | mammary gland branching involved in thelarche | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 2 | 6 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 3 | 6 |
| GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 2 | 6 |
| GO:0042981 | regulation of apoptotic process | 4.58e-03 | 1.00e+00 | 4.265 | 2 | 4 | 150 |
| GO:0006974 | cellular response to DNA damage stimulus | 4.64e-03 | 1.00e+00 | 4.256 | 2 | 7 | 151 |
| GO:0008347 | glial cell migration | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0071354 | cellular response to interleukin-6 | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0001955 | blood vessel maturation | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0003016 | respiratory system process | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0006983 | ER overload response | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0003714 | transcription corepressor activity | 6.39e-03 | 1.00e+00 | 4.018 | 2 | 11 | 178 |
| GO:0035413 | positive regulation of catenin import into nucleus | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 2 | 10 |
| GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 2 | 10 |
| GO:0003677 | DNA binding | 7.02e-03 | 1.00e+00 | 2.244 | 4 | 49 | 1218 |
| GO:0045662 | negative regulation of myoblast differentiation | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 3 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 4 | 11 |
| GO:0005838 | proteasome regulatory particle | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0035198 | miRNA binding | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 3 | 13 |
| GO:0007413 | axonal fasciculation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0071480 | cellular response to gamma radiation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0030234 | enzyme regulator activity | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 13 |
| GO:0010467 | gene expression | 9.33e-03 | 1.00e+00 | 2.693 | 3 | 45 | 669 |
| GO:0001824 | blastocyst development | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 15 |
| GO:0060749 | mammary gland alveolus development | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 15 |
| GO:0042176 | regulation of protein catabolic process | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 2 | 16 |
| GO:0008134 | transcription factor binding | 1.15e-02 | 1.00e+00 | 3.575 | 2 | 18 | 242 |
| GO:0022624 | proteasome accessory complex | 1.17e-02 | 1.00e+00 | 6.407 | 1 | 1 | 17 |
| GO:0043015 | gamma-tubulin binding | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 18 |
| GO:0032757 | positive regulation of interleukin-8 production | 1.38e-02 | 1.00e+00 | 6.172 | 1 | 1 | 20 |
| GO:0005634 | nucleus | 1.44e-02 | 1.00e+00 | 1.147 | 7 | 159 | 4559 |
| GO:0048675 | axon extension | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 2 | 21 |
| GO:0016575 | histone deacetylation | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 3 | 21 |
| GO:0005743 | mitochondrial inner membrane | 1.49e-02 | 1.00e+00 | 3.380 | 2 | 6 | 277 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 2 | 22 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 1 | 23 |
| GO:0035329 | hippo signaling | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 5 | 26 |
| GO:0051209 | release of sequestered calcium ion into cytosol | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 26 |
| GO:0005829 | cytosol | 1.81e-02 | 1.00e+00 | 1.531 | 5 | 86 | 2496 |
| GO:0030331 | estrogen receptor binding | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 2 | 27 |
| GO:0071897 | DNA biosynthetic process | 1.93e-02 | 1.00e+00 | 5.687 | 1 | 1 | 28 |
| GO:0042594 | response to starvation | 2.13e-02 | 1.00e+00 | 5.540 | 1 | 4 | 31 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 2.20e-02 | 1.00e+00 | 5.494 | 1 | 2 | 32 |
| GO:0043565 | sequence-specific DNA binding | 2.27e-02 | 1.00e+00 | 3.060 | 2 | 11 | 346 |
| GO:0008285 | negative regulation of cell proliferation | 2.36e-02 | 1.00e+00 | 3.031 | 2 | 10 | 353 |
| GO:0035019 | somatic stem cell maintenance | 2.74e-02 | 1.00e+00 | 5.172 | 1 | 6 | 40 |
| GO:0043525 | positive regulation of neuron apoptotic process | 3.14e-02 | 1.00e+00 | 4.971 | 1 | 4 | 46 |
| GO:0019233 | sensory perception of pain | 3.28e-02 | 1.00e+00 | 4.909 | 1 | 1 | 48 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 1 | 50 |
| GO:0006986 | response to unfolded protein | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 4 | 50 |
| GO:0040008 | regulation of growth | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 2 | 50 |
| GO:0034976 | response to endoplasmic reticulum stress | 3.48e-02 | 1.00e+00 | 4.822 | 1 | 2 | 51 |
| GO:0045454 | cell redox homeostasis | 3.48e-02 | 1.00e+00 | 4.822 | 1 | 1 | 51 |
| GO:0009611 | response to wounding | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 5 | 52 |
| GO:0043204 | perikaryon | 3.75e-02 | 1.00e+00 | 4.713 | 1 | 1 | 55 |
| GO:0000502 | proteasome complex | 3.95e-02 | 1.00e+00 | 4.636 | 1 | 3 | 58 |
| GO:0006396 | RNA processing | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 5 | 59 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 6 | 59 |
| GO:0042995 | cell projection | 4.08e-02 | 1.00e+00 | 4.587 | 1 | 3 | 60 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 4.14e-02 | 1.00e+00 | 2.593 | 2 | 24 | 478 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 4.15e-02 | 1.00e+00 | 4.563 | 1 | 2 | 61 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 4.35e-02 | 1.00e+00 | 4.494 | 1 | 1 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 2 | 65 |
| GO:0010468 | regulation of gene expression | 4.62e-02 | 1.00e+00 | 4.407 | 1 | 2 | 68 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.75e-02 | 1.00e+00 | 4.365 | 1 | 2 | 70 |
| GO:0042826 | histone deacetylase binding | 4.88e-02 | 1.00e+00 | 4.324 | 1 | 4 | 72 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 4.88e-02 | 1.00e+00 | 4.324 | 1 | 1 | 72 |
| GO:0009791 | post-embryonic development | 4.95e-02 | 1.00e+00 | 4.304 | 1 | 3 | 73 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 4 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 2 | 74 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 1 | 75 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 1 | 75 |
| GO:0071013 | catalytic step 2 spliceosome | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 7 | 78 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 5.35e-02 | 1.00e+00 | 4.190 | 1 | 2 | 79 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 5.35e-02 | 1.00e+00 | 4.190 | 1 | 2 | 79 |
| GO:0009653 | anatomical structure morphogenesis | 5.61e-02 | 1.00e+00 | 4.119 | 1 | 2 | 83 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 5.81e-02 | 1.00e+00 | 4.068 | 1 | 8 | 86 |
| GO:0042470 | melanosome | 6.14e-02 | 1.00e+00 | 3.986 | 1 | 2 | 91 |
| GO:0016363 | nuclear matrix | 6.14e-02 | 1.00e+00 | 3.986 | 1 | 6 | 91 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 6.20e-02 | 1.00e+00 | 3.971 | 1 | 1 | 92 |
| GO:0001764 | neuron migration | 6.27e-02 | 1.00e+00 | 3.955 | 1 | 1 | 93 |
| GO:0005770 | late endosome | 6.27e-02 | 1.00e+00 | 3.955 | 1 | 2 | 93 |
| GO:0007417 | central nervous system development | 6.46e-02 | 1.00e+00 | 3.909 | 1 | 5 | 96 |
| GO:0005515 | protein binding | 6.90e-02 | 1.00e+00 | 0.745 | 7 | 198 | 6024 |
| GO:0070062 | extracellular vesicular exosome | 6.93e-02 | 1.00e+00 | 1.265 | 4 | 57 | 2400 |
| GO:0030308 | negative regulation of cell growth | 7.37e-02 | 1.00e+00 | 3.713 | 1 | 6 | 110 |
| GO:0030529 | ribonucleoprotein complex | 7.50e-02 | 1.00e+00 | 3.687 | 1 | 5 | 112 |
| GO:0000209 | protein polyubiquitination | 7.76e-02 | 1.00e+00 | 3.636 | 1 | 3 | 116 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 8.15e-02 | 1.00e+00 | 2.045 | 2 | 39 | 699 |
| GO:0007050 | cell cycle arrest | 8.27e-02 | 1.00e+00 | 3.540 | 1 | 5 | 124 |
| GO:0016055 | Wnt signaling pathway | 9.17e-02 | 1.00e+00 | 3.386 | 1 | 4 | 138 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 9.87e-02 | 1.00e+00 | 3.275 | 1 | 3 | 149 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 1.00e-01 | 1.00e+00 | 1.870 | 2 | 41 | 789 |
| GO:0016020 | membrane | 1.01e-01 | 1.00e+00 | 1.364 | 3 | 46 | 1681 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.08e-01 | 1.00e+00 | 3.137 | 1 | 15 | 164 |
| GO:0034641 | cellular nitrogen compound metabolic process | 1.12e-01 | 1.00e+00 | 3.076 | 1 | 2 | 171 |
| GO:0005667 | transcription factor complex | 1.15e-01 | 1.00e+00 | 3.043 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.17e-01 | 1.00e+00 | 3.018 | 1 | 5 | 178 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.21e-01 | 1.00e+00 | 2.971 | 1 | 22 | 184 |
| GO:0016071 | mRNA metabolic process | 1.44e-01 | 1.00e+00 | 2.693 | 1 | 8 | 223 |
| GO:0008380 | RNA splicing | 1.47e-01 | 1.00e+00 | 2.661 | 1 | 22 | 228 |
| GO:0007399 | nervous system development | 1.49e-01 | 1.00e+00 | 2.642 | 1 | 6 | 231 |
| GO:0005739 | mitochondrion | 1.49e-01 | 1.00e+00 | 1.531 | 2 | 23 | 998 |
| GO:0003713 | transcription coactivator activity | 1.53e-01 | 1.00e+00 | 2.605 | 1 | 24 | 237 |
| GO:0016070 | RNA metabolic process | 1.59e-01 | 1.00e+00 | 2.546 | 1 | 8 | 247 |
| GO:0044822 | poly(A) RNA binding | 1.63e-01 | 1.00e+00 | 1.450 | 2 | 50 | 1056 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 1.72e-01 | 1.00e+00 | 2.423 | 1 | 7 | 269 |
| GO:0019899 | enzyme binding | 1.76e-01 | 1.00e+00 | 2.380 | 1 | 9 | 277 |
| GO:0043234 | protein complex | 1.87e-01 | 1.00e+00 | 2.290 | 1 | 11 | 295 |
| GO:0005813 | centrosome | 2.04e-01 | 1.00e+00 | 2.145 | 1 | 7 | 326 |
| GO:0008283 | cell proliferation | 2.06e-01 | 1.00e+00 | 2.137 | 1 | 14 | 328 |
| GO:0003723 | RNA binding | 2.13e-01 | 1.00e+00 | 2.076 | 1 | 18 | 342 |
| GO:0046982 | protein heterodimerization activity | 2.34e-01 | 1.00e+00 | 1.924 | 1 | 8 | 380 |
| GO:0008150 | biological_process | 2.39e-01 | 1.00e+00 | 1.891 | 1 | 4 | 389 |
| GO:0000278 | mitotic cell cycle | 2.40e-01 | 1.00e+00 | 1.883 | 1 | 15 | 391 |
| GO:0009986 | cell surface | 2.46e-01 | 1.00e+00 | 1.843 | 1 | 9 | 402 |
| GO:0005737 | cytoplasm | 2.51e-01 | 1.00e+00 | 0.615 | 4 | 124 | 3767 |
| GO:0006366 | transcription from RNA polymerase II promoter | 2.55e-01 | 1.00e+00 | 1.783 | 1 | 30 | 419 |
| GO:0043066 | negative regulation of apoptotic process | 2.58e-01 | 1.00e+00 | 1.766 | 1 | 16 | 424 |
| GO:0006351 | transcription, DNA-templated | 2.65e-01 | 1.00e+00 | 0.996 | 2 | 57 | 1446 |
| GO:0044267 | cellular protein metabolic process | 2.84e-01 | 1.00e+00 | 1.605 | 1 | 20 | 474 |
| GO:0016032 | viral process | 3.14e-01 | 1.00e+00 | 1.433 | 1 | 26 | 534 |
| GO:0006915 | apoptotic process | 3.25e-01 | 1.00e+00 | 1.378 | 1 | 12 | 555 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.32e-01 | 1.00e+00 | 1.339 | 1 | 37 | 570 |
| GO:0005887 | integral component of plasma membrane | 4.70e-01 | 1.00e+00 | 0.700 | 1 | 10 | 888 |
| GO:0007165 | signal transduction | 4.78e-01 | 1.00e+00 | 0.669 | 1 | 13 | 907 |
| GO:0044281 | small molecule metabolic process | 5.84e-01 | 1.00e+00 | 0.252 | 1 | 20 | 1211 |
| GO:0005524 | ATP binding | 6.11e-01 | 1.00e+00 | 0.152 | 1 | 31 | 1298 |
| GO:0016021 | integral component of membrane | 7.72e-01 | 1.00e+00 | -0.459 | 1 | 17 | 1982 |