int-snw-1432
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.955 | 8.09e-17 | 1.07e-04 | 1.17e-03 |
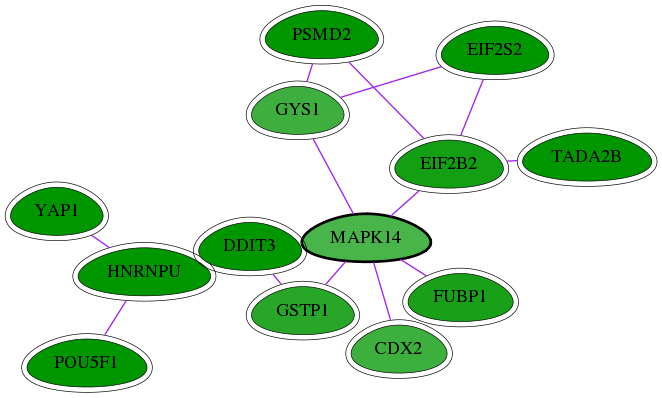
Genes (13)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| PSMD2 | 5708 | 47 | -4.172 | 3.157 | 386 | Yes | Yes |
| EIF2S2 | 8894 | 9 | -2.999 | 2.968 | 81 | Yes | Yes |
| TADA2B | 93624 | 11 | -3.061 | 3.082 | 19 | Yes | - |
| GYS1 | 2997 | 1 | -2.013 | 2.955 | 88 | Yes | - |
| EIF2B2 | 8892 | 8 | -2.462 | 2.968 | 95 | Yes | Yes |
| YAP1 | 10413 | 94 | -4.256 | 3.538 | 53 | Yes | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| CDX2 | 1045 | 3 | -2.037 | 2.955 | 33 | Yes | Yes |
| HNRNPU | 3192 | 76 | -2.869 | 3.538 | 139 | Yes | - |
| [ MAPK14 ] | 1432 | 4 | -1.902 | 2.955 | 272 | - | - |
| GSTP1 | 2950 | 3 | -2.251 | 2.955 | 59 | Yes | - |
| DDIT3 | 1649 | 60 | -2.825 | 3.504 | 343 | Yes | Yes |
| FUBP1 | 8880 | 3 | -2.414 | 2.955 | 23 | Yes | Yes |
Interactions (15)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| GYS1 | 2997 | EIF2S2 | 8894 | pp | -- | int.I2D: YeastLow |
| GYS1 | 2997 | PSMD2 | 5708 | pp | -- | int.I2D: YeastLow |
| EIF2B2 | 8892 | EIF2S2 | 8894 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh |
| EIF2B2 | 8892 | TADA2B | 93624 | pp | -- | int.I2D: BioGrid_Fly, BIND_Fly, FlyLow, IntAct_Fly, MINT_Fly |
| MAPK14 | 1432 | GSTP1 | 2950 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, BCI |
| HNRNPU | 3192 | YAP1 | 10413 | pp | -- | int.I2D: HPRD; int.HPRD: in vitro, in vivo |
| MAPK14 | 1432 | GYS1 | 2997 | pp | -- | int.I2D: IntAct_Worm, BioGrid_Worm, BIND_Worm, CORE_1, MINT_Worm |
| DDIT3 | 1649 | HNRNPU | 3192 | pp | -- | int.I2D: BEHRENDS_AUTOPHAGY_LOW |
| DDIT3 | 1649 | GSTP1 | 2950 | pp | -- | int.I2D: BEHRENDS_AUTOPHAGY_LOW |
| CDX2 | 1045 | MAPK14 | 1432 | pp | -- | int.I2D: BCI, HPRD; int.HPRD: in vitro |
| MAPK14 | 1432 | DDIT3 | 1649 | pp | -- | int.I2D: BCI, BioGrid, HPRD; int.HPRD: in vitro |
| HNRNPU | 3192 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| MAPK14 | 1432 | EIF2B2 | 8892 | pp | -- | int.I2D: IntAct, SOURAV_MAPK_LOW |
| PSMD2 | 5708 | EIF2B2 | 8892 | pp | -- | int.I2D: IntAct_Yeast |
| MAPK14 | 1432 | FUBP1 | 8880 | pp | -- | int.I2D: BioGrid |
Related GO terms (266)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0010467 | gene expression | 1.26e-05 | 1.82e-01 | 3.315 | 6 | 45 | 669 |
| GO:0005829 | cytosol | 5.02e-05 | 7.24e-01 | 2.000 | 9 | 86 | 2496 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 5.82e-05 | 8.39e-01 | 7.415 | 2 | 2 | 13 |
| GO:0005654 | nucleoplasm | 1.90e-04 | 1.00e+00 | 2.621 | 6 | 64 | 1082 |
| GO:0044212 | transcription regulatory region DNA binding | 4.07e-04 | 1.00e+00 | 4.308 | 3 | 17 | 168 |
| GO:0003714 | transcription corepressor activity | 4.83e-04 | 1.00e+00 | 4.225 | 3 | 11 | 178 |
| GO:0035019 | somatic stem cell maintenance | 5.74e-04 | 1.00e+00 | 5.794 | 2 | 6 | 40 |
| GO:0003743 | translation initiation factor activity | 8.61e-04 | 1.00e+00 | 5.501 | 2 | 7 | 49 |
| GO:0070664 | negative regulation of leukocyte proliferation | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0035732 | nitric oxide storage | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:2000016 | negative regulation of determination of dorsal identity | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0097057 | TRAF2-GSTP1 complex | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0070026 | nitric oxide binding | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0035731 | dinitrosyl-iron complex binding | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0038066 | p38MAPK cascade | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0035730 | S-nitrosoglutathione binding | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0060242 | contact inhibition | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0009890 | negative regulation of biosynthetic process | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0002176 | male germ cell proliferation | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0019207 | kinase regulator activity | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0061547 | glycogen synthase activity, transferring glucose-1-phosphate | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0004373 | glycogen (starch) synthase activity | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0044324 | regulation of transcription involved in anterior/posterior axis specification | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0005515 | protein binding | 2.00e-03 | 1.00e+00 | 1.019 | 11 | 198 | 6024 |
| GO:0071222 | cellular response to lipopolysaccharide | 2.33e-03 | 1.00e+00 | 4.776 | 2 | 4 | 81 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2.63e-03 | 1.00e+00 | 4.689 | 2 | 8 | 86 |
| GO:0035726 | common myeloid progenitor cell proliferation | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0090400 | stress-induced premature senescence | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 2.75e-03 | 1.00e+00 | 2.666 | 4 | 39 | 699 |
| GO:0007417 | central nervous system development | 3.26e-03 | 1.00e+00 | 4.531 | 2 | 5 | 96 |
| GO:0043565 | sequence-specific DNA binding | 3.27e-03 | 1.00e+00 | 3.266 | 3 | 11 | 346 |
| GO:0051716 | cellular response to stimulus | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 4 | 4 |
| GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0046523 | S-methyl-5-thioribose-1-phosphate isomerase activity | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 3 | 4 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 4.27e-03 | 1.00e+00 | 2.492 | 4 | 41 | 789 |
| GO:0070461 | SAGA-type complex | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 5 |
| GO:0001714 | endodermal cell fate specification | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0043620 | regulation of DNA-templated transcription in response to stress | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 5 |
| GO:0051525 | NFAT protein binding | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 5 |
| GO:1901741 | positive regulation of myoblast fusion | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0006006 | glucose metabolic process | 4.88e-03 | 1.00e+00 | 4.233 | 2 | 5 | 118 |
| GO:0032873 | negative regulation of stress-activated MAPK cascade | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 2 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 3 | 6 |
| GO:0032691 | negative regulation of interleukin-1 beta production | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 1 | 6 |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 4 | 6 |
| GO:0008432 | JUN kinase binding | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 1 | 6 |
| GO:0006366 | transcription from RNA polymerase II promoter | 5.60e-03 | 1.00e+00 | 2.990 | 3 | 30 | 419 |
| GO:0006413 | translational initiation | 5.98e-03 | 1.00e+00 | 4.082 | 2 | 7 | 131 |
| GO:0032930 | positive regulation of superoxide anion generation | 6.29e-03 | 1.00e+00 | 7.308 | 1 | 2 | 7 |
| GO:0002674 | negative regulation of acute inflammatory response | 6.29e-03 | 1.00e+00 | 7.308 | 1 | 1 | 7 |
| GO:0032872 | regulation of stress-activated MAPK cascade | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0001955 | blood vessel maturation | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0005536 | glucose binding | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0010831 | positive regulation of myotube differentiation | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0006974 | cellular response to DNA damage stimulus | 7.87e-03 | 1.00e+00 | 3.877 | 2 | 7 | 151 |
| GO:0044267 | cellular protein metabolic process | 7.89e-03 | 1.00e+00 | 2.812 | 3 | 20 | 474 |
| GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0000302 | response to reactive oxygen species | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0019509 | L-methionine biosynthetic process from methylthioadenosine | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 3 | 9 |
| GO:0006983 | ER overload response | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0060711 | labyrinthine layer development | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 3 | 9 |
| GO:0032495 | response to muramyl dipeptide | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 2 | 9 |
| GO:0035413 | positive regulation of catenin import into nucleus | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 10 |
| GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 10 |
| GO:0006355 | regulation of transcription, DNA-templated | 9.80e-03 | 1.00e+00 | 2.154 | 4 | 43 | 997 |
| GO:0045663 | positive regulation of myoblast differentiation | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 2 | 11 |
| GO:0045662 | negative regulation of myoblast differentiation | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 3 | 11 |
| GO:0019395 | fatty acid oxidation | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 2 | 11 |
| GO:0004707 | MAP kinase activity | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 11 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 11 |
| GO:0042770 | signal transduction in response to DNA damage | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 4 | 11 |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 3 | 12 |
| GO:0051146 | striated muscle cell differentiation | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 12 |
| GO:0005838 | proteasome regulatory particle | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 12 |
| GO:0035198 | miRNA binding | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 12 |
| GO:0043508 | negative regulation of JUN kinase activity | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0071480 | cellular response to gamma radiation | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0004708 | MAP kinase kinase activity | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0030914 | STAGA complex | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 3 | 13 |
| GO:0043409 | negative regulation of MAPK cascade | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 2 | 13 |
| GO:0030234 | enzyme regulator activity | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 2 | 13 |
| GO:0044822 | poly(A) RNA binding | 1.20e-02 | 1.00e+00 | 2.071 | 4 | 50 | 1056 |
| GO:0042307 | positive regulation of protein import into nucleus | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 4 | 15 |
| GO:0001824 | blastocyst development | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 15 |
| GO:0042176 | regulation of protein catabolic process | 1.43e-02 | 1.00e+00 | 6.116 | 1 | 2 | 16 |
| GO:0001829 | trophectodermal cell differentiation | 1.52e-02 | 1.00e+00 | 6.028 | 1 | 2 | 17 |
| GO:0070372 | regulation of ERK1 and ERK2 cascade | 1.52e-02 | 1.00e+00 | 6.028 | 1 | 2 | 17 |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1.52e-02 | 1.00e+00 | 6.028 | 1 | 3 | 17 |
| GO:0022624 | proteasome accessory complex | 1.52e-02 | 1.00e+00 | 6.028 | 1 | 1 | 17 |
| GO:0004364 | glutathione transferase activity | 1.61e-02 | 1.00e+00 | 5.946 | 1 | 1 | 18 |
| GO:0016071 | mRNA metabolic process | 1.66e-02 | 1.00e+00 | 3.315 | 2 | 8 | 223 |
| GO:0016234 | inclusion body | 1.70e-02 | 1.00e+00 | 5.868 | 1 | 2 | 19 |
| GO:0006412 | translation | 1.76e-02 | 1.00e+00 | 3.270 | 2 | 8 | 230 |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:0001502 | cartilage condensation | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:0032757 | positive regulation of interleukin-8 production | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:1901687 | glutathione derivative biosynthetic process | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 3 | 20 |
| GO:0014003 | oligodendrocyte development | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 5 | 20 |
| GO:0045648 | positive regulation of erythrocyte differentiation | 1.88e-02 | 1.00e+00 | 5.723 | 1 | 1 | 21 |
| GO:0008134 | transcription factor binding | 1.94e-02 | 1.00e+00 | 3.197 | 2 | 18 | 242 |
| GO:0003677 | DNA binding | 1.95e-02 | 1.00e+00 | 1.865 | 4 | 49 | 1218 |
| GO:0030316 | osteoclast differentiation | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 2 | 22 |
| GO:0048147 | negative regulation of fibroblast proliferation | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 1 | 22 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 2 | 22 |
| GO:0001205 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 4 | 22 |
| GO:0016070 | RNA metabolic process | 2.01e-02 | 1.00e+00 | 3.167 | 2 | 8 | 247 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2.05e-02 | 1.00e+00 | 5.592 | 1 | 1 | 23 |
| GO:0071479 | cellular response to ionizing radiation | 2.05e-02 | 1.00e+00 | 5.592 | 1 | 1 | 23 |
| GO:2000379 | positive regulation of reactive oxygen species metabolic process | 2.05e-02 | 1.00e+00 | 5.592 | 1 | 2 | 23 |
| GO:0008135 | translation factor activity, nucleic acid binding | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 3 | 24 |
| GO:0000794 | condensed nuclear chromosome | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 1 | 24 |
| GO:0005978 | glycogen biosynthetic process | 2.32e-02 | 1.00e+00 | 5.415 | 1 | 1 | 26 |
| GO:0035329 | hippo signaling | 2.32e-02 | 1.00e+00 | 5.415 | 1 | 5 | 26 |
| GO:0051209 | release of sequestered calcium ion into cytosol | 2.32e-02 | 1.00e+00 | 5.415 | 1 | 1 | 26 |
| GO:0051149 | positive regulation of muscle cell differentiation | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 4 | 27 |
| GO:0032720 | negative regulation of tumor necrosis factor production | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 2 | 27 |
| GO:0000077 | DNA damage checkpoint | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 1 | 27 |
| GO:0045597 | positive regulation of cell differentiation | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 1 | 27 |
| GO:0006749 | glutathione metabolic process | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 1 | 27 |
| GO:0005634 | nucleus | 2.51e-02 | 1.00e+00 | 0.961 | 8 | 159 | 4559 |
| GO:0008333 | endosome to lysosome transport | 2.58e-02 | 1.00e+00 | 5.258 | 1 | 2 | 29 |
| GO:0031663 | lipopolysaccharide-mediated signaling pathway | 2.58e-02 | 1.00e+00 | 5.258 | 1 | 2 | 29 |
| GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 2 | 30 |
| GO:0042594 | response to starvation | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 4 | 31 |
| GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 1 | 31 |
| GO:0043407 | negative regulation of MAP kinase activity | 2.85e-02 | 1.00e+00 | 5.116 | 1 | 1 | 32 |
| GO:0002062 | chondrocyte differentiation | 2.85e-02 | 1.00e+00 | 5.116 | 1 | 2 | 32 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 2.85e-02 | 1.00e+00 | 5.116 | 1 | 2 | 32 |
| GO:0042692 | muscle cell differentiation | 3.02e-02 | 1.00e+00 | 5.028 | 1 | 4 | 34 |
| GO:0001568 | blood vessel development | 3.11e-02 | 1.00e+00 | 4.986 | 1 | 2 | 35 |
| GO:0042552 | myelination | 3.11e-02 | 1.00e+00 | 4.986 | 1 | 4 | 35 |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 3.11e-02 | 1.00e+00 | 4.986 | 1 | 2 | 35 |
| GO:0006446 | regulation of translational initiation | 3.20e-02 | 1.00e+00 | 4.946 | 1 | 4 | 36 |
| GO:0001541 | ovarian follicle development | 3.29e-02 | 1.00e+00 | 4.906 | 1 | 2 | 37 |
| GO:0009408 | response to heat | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 6 | 40 |
| GO:0007519 | skeletal muscle tissue development | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 2 | 40 |
| GO:0003723 | RNA binding | 3.68e-02 | 1.00e+00 | 2.698 | 2 | 18 | 342 |
| GO:0043434 | response to peptide hormone | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 5 | 45 |
| GO:0043525 | positive regulation of neuron apoptotic process | 4.07e-02 | 1.00e+00 | 4.592 | 1 | 4 | 46 |
| GO:0000902 | cell morphogenesis | 4.33e-02 | 1.00e+00 | 4.501 | 1 | 4 | 49 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 1 | 50 |
| GO:0006986 | response to unfolded protein | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 4 | 50 |
| GO:0034976 | response to endoplasmic reticulum stress | 4.50e-02 | 1.00e+00 | 4.443 | 1 | 2 | 51 |
| GO:0045454 | cell redox homeostasis | 4.50e-02 | 1.00e+00 | 4.443 | 1 | 1 | 51 |
| GO:0017053 | transcriptional repressor complex | 4.50e-02 | 1.00e+00 | 4.443 | 1 | 2 | 51 |
| GO:0009611 | response to wounding | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 5 | 52 |
| GO:0051403 | stress-activated MAPK cascade | 4.76e-02 | 1.00e+00 | 4.361 | 1 | 2 | 54 |
| GO:0000502 | proteasome complex | 5.11e-02 | 1.00e+00 | 4.258 | 1 | 3 | 58 |
| GO:0006396 | RNA processing | 5.19e-02 | 1.00e+00 | 4.233 | 1 | 5 | 59 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 5.19e-02 | 1.00e+00 | 4.233 | 1 | 6 | 59 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 5.36e-02 | 1.00e+00 | 4.185 | 1 | 2 | 61 |
| GO:0043066 | negative regulation of apoptotic process | 5.43e-02 | 1.00e+00 | 2.388 | 2 | 16 | 424 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 5.62e-02 | 1.00e+00 | 4.116 | 1 | 1 | 64 |
| GO:0009749 | response to glucose | 5.62e-02 | 1.00e+00 | 4.116 | 1 | 7 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 2 | 65 |
| GO:0034166 | toll-like receptor 10 signaling pathway | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 4 | 65 |
| GO:0006469 | negative regulation of protein kinase activity | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 4 | 65 |
| GO:0034146 | toll-like receptor 5 signaling pathway | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 4 | 65 |
| GO:0003697 | single-stranded DNA binding | 5.96e-02 | 1.00e+00 | 4.028 | 1 | 5 | 68 |
| GO:0010468 | regulation of gene expression | 5.96e-02 | 1.00e+00 | 4.028 | 1 | 2 | 68 |
| GO:0018105 | peptidyl-serine phosphorylation | 6.05e-02 | 1.00e+00 | 4.007 | 1 | 2 | 69 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 6.13e-02 | 1.00e+00 | 3.986 | 1 | 2 | 70 |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 6.22e-02 | 1.00e+00 | 3.966 | 1 | 5 | 71 |
| GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway | 6.22e-02 | 1.00e+00 | 3.966 | 1 | 4 | 71 |
| GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway | 6.22e-02 | 1.00e+00 | 3.966 | 1 | 4 | 71 |
| GO:0034162 | toll-like receptor 9 signaling pathway | 6.30e-02 | 1.00e+00 | 3.946 | 1 | 4 | 72 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 6.30e-02 | 1.00e+00 | 3.946 | 1 | 1 | 72 |
| GO:0034134 | toll-like receptor 2 signaling pathway | 6.39e-02 | 1.00e+00 | 3.926 | 1 | 4 | 73 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 6.47e-02 | 1.00e+00 | 3.906 | 1 | 4 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 6.47e-02 | 1.00e+00 | 3.906 | 1 | 2 | 74 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 6.56e-02 | 1.00e+00 | 3.887 | 1 | 1 | 75 |
| GO:0008584 | male gonad development | 6.56e-02 | 1.00e+00 | 3.887 | 1 | 3 | 75 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 6.56e-02 | 1.00e+00 | 3.887 | 1 | 1 | 75 |
| GO:0007265 | Ras protein signal transduction | 6.56e-02 | 1.00e+00 | 3.887 | 1 | 3 | 75 |
| GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 6.64e-02 | 1.00e+00 | 3.868 | 1 | 3 | 76 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 6.72e-02 | 1.00e+00 | 2.215 | 2 | 24 | 478 |
| GO:0071013 | catalytic step 2 spliceosome | 6.81e-02 | 1.00e+00 | 3.830 | 1 | 7 | 78 |
| GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 6.81e-02 | 1.00e+00 | 3.830 | 1 | 3 | 78 |
| GO:0034138 | toll-like receptor 3 signaling pathway | 6.89e-02 | 1.00e+00 | 3.812 | 1 | 3 | 79 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 6.89e-02 | 1.00e+00 | 3.812 | 1 | 2 | 79 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 6.89e-02 | 1.00e+00 | 3.812 | 1 | 2 | 79 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 6.98e-02 | 1.00e+00 | 3.794 | 1 | 5 | 80 |
| GO:0009653 | anatomical structure morphogenesis | 7.23e-02 | 1.00e+00 | 3.741 | 1 | 2 | 83 |
| GO:0009952 | anterior/posterior pattern specification | 7.23e-02 | 1.00e+00 | 3.741 | 1 | 4 | 83 |
| GO:0009887 | organ morphogenesis | 7.48e-02 | 1.00e+00 | 3.689 | 1 | 5 | 86 |
| GO:0003690 | double-stranded DNA binding | 7.48e-02 | 1.00e+00 | 3.689 | 1 | 3 | 86 |
| GO:0000922 | spindle pole | 7.57e-02 | 1.00e+00 | 3.673 | 1 | 4 | 87 |
| GO:0000187 | activation of MAPK activity | 7.82e-02 | 1.00e+00 | 3.624 | 1 | 3 | 90 |
| GO:0006928 | cellular component movement | 7.90e-02 | 1.00e+00 | 3.608 | 1 | 3 | 91 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 7.99e-02 | 1.00e+00 | 3.592 | 1 | 1 | 92 |
| GO:0001649 | osteoblast differentiation | 7.99e-02 | 1.00e+00 | 3.592 | 1 | 5 | 92 |
| GO:0005770 | late endosome | 8.07e-02 | 1.00e+00 | 3.576 | 1 | 2 | 93 |
| GO:0034142 | toll-like receptor 4 signaling pathway | 8.32e-02 | 1.00e+00 | 3.531 | 1 | 4 | 96 |
| GO:0006915 | apoptotic process | 8.71e-02 | 1.00e+00 | 1.999 | 2 | 12 | 555 |
| GO:0006805 | xenobiotic metabolic process | 8.90e-02 | 1.00e+00 | 3.429 | 1 | 2 | 103 |
| GO:0044281 | small molecule metabolic process | 8.95e-02 | 1.00e+00 | 1.459 | 3 | 20 | 1211 |
| GO:0006935 | chemotaxis | 9.07e-02 | 1.00e+00 | 3.401 | 1 | 3 | 105 |
| GO:0002224 | toll-like receptor signaling pathway | 9.39e-02 | 1.00e+00 | 3.347 | 1 | 4 | 109 |
| GO:0030529 | ribonucleoprotein complex | 9.64e-02 | 1.00e+00 | 3.308 | 1 | 5 | 112 |
| GO:0000209 | protein polyubiquitination | 9.97e-02 | 1.00e+00 | 3.258 | 1 | 3 | 116 |
| GO:0006325 | chromatin organization | 1.01e-01 | 1.00e+00 | 3.233 | 1 | 12 | 118 |
| GO:0005524 | ATP binding | 1.05e-01 | 1.00e+00 | 1.359 | 3 | 31 | 1298 |
| GO:0007050 | cell cycle arrest | 1.06e-01 | 1.00e+00 | 3.161 | 1 | 5 | 124 |
| GO:0031982 | vesicle | 1.12e-01 | 1.00e+00 | 3.082 | 1 | 3 | 131 |
| GO:0016055 | Wnt signaling pathway | 1.18e-01 | 1.00e+00 | 3.007 | 1 | 4 | 138 |
| GO:0007507 | heart development | 1.18e-01 | 1.00e+00 | 2.997 | 1 | 7 | 139 |
| GO:0010628 | positive regulation of gene expression | 1.24e-01 | 1.00e+00 | 2.926 | 1 | 7 | 146 |
| GO:0007166 | cell surface receptor signaling pathway | 1.25e-01 | 1.00e+00 | 2.916 | 1 | 4 | 147 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.26e-01 | 1.00e+00 | 2.896 | 1 | 3 | 149 |
| GO:0042981 | regulation of apoptotic process | 1.27e-01 | 1.00e+00 | 2.887 | 1 | 4 | 150 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.38e-01 | 1.00e+00 | 2.758 | 1 | 15 | 164 |
| GO:0034641 | cellular nitrogen compound metabolic process | 1.44e-01 | 1.00e+00 | 2.698 | 1 | 2 | 171 |
| GO:0005667 | transcription factor complex | 1.47e-01 | 1.00e+00 | 2.664 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.49e-01 | 1.00e+00 | 2.640 | 1 | 5 | 178 |
| GO:0043547 | positive regulation of GTPase activity | 1.51e-01 | 1.00e+00 | 2.624 | 1 | 9 | 180 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.54e-01 | 1.00e+00 | 2.592 | 1 | 22 | 184 |
| GO:0001525 | angiogenesis | 1.59e-01 | 1.00e+00 | 2.538 | 1 | 7 | 191 |
| GO:0001701 | in utero embryonic development | 1.67e-01 | 1.00e+00 | 2.465 | 1 | 8 | 201 |
| GO:0030168 | platelet activation | 1.70e-01 | 1.00e+00 | 2.436 | 1 | 6 | 205 |
| GO:0005622 | intracellular | 1.71e-01 | 1.00e+00 | 2.422 | 1 | 6 | 207 |
| GO:0016020 | membrane | 1.87e-01 | 1.00e+00 | 0.986 | 3 | 46 | 1681 |
| GO:0008380 | RNA splicing | 1.87e-01 | 1.00e+00 | 2.283 | 1 | 22 | 228 |
| GO:0003713 | transcription coactivator activity | 1.94e-01 | 1.00e+00 | 2.227 | 1 | 24 | 237 |
| GO:0005975 | carbohydrate metabolic process | 2.06e-01 | 1.00e+00 | 2.133 | 1 | 8 | 253 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 2.14e-01 | 1.00e+00 | 2.071 | 1 | 23 | 264 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 2.17e-01 | 1.00e+00 | 2.044 | 1 | 7 | 269 |
| GO:0005739 | mitochondrion | 2.26e-01 | 1.00e+00 | 1.153 | 2 | 23 | 998 |
| GO:0005737 | cytoplasm | 2.35e-01 | 1.00e+00 | 0.558 | 5 | 124 | 3767 |
| GO:0035556 | intracellular signal transduction | 2.41e-01 | 1.00e+00 | 1.872 | 1 | 9 | 303 |
| GO:0004674 | protein serine/threonine kinase activity | 2.45e-01 | 1.00e+00 | 1.849 | 1 | 6 | 308 |
| GO:0005525 | GTP binding | 2.46e-01 | 1.00e+00 | 1.840 | 1 | 12 | 310 |
| GO:0019901 | protein kinase binding | 2.51e-01 | 1.00e+00 | 1.807 | 1 | 18 | 317 |
| GO:0003682 | chromatin binding | 2.58e-01 | 1.00e+00 | 1.762 | 1 | 19 | 327 |
| GO:0008283 | cell proliferation | 2.59e-01 | 1.00e+00 | 1.758 | 1 | 14 | 328 |
| GO:0046982 | protein heterodimerization activity | 2.93e-01 | 1.00e+00 | 1.546 | 1 | 8 | 380 |
| GO:0008284 | positive regulation of cell proliferation | 2.95e-01 | 1.00e+00 | 1.538 | 1 | 10 | 382 |
| GO:0000278 | mitotic cell cycle | 3.01e-01 | 1.00e+00 | 1.505 | 1 | 15 | 391 |
| GO:0009986 | cell surface | 3.08e-01 | 1.00e+00 | 1.465 | 1 | 9 | 402 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.15e-01 | 1.00e+00 | 1.426 | 1 | 24 | 413 |
| GO:0007596 | blood coagulation | 3.41e-01 | 1.00e+00 | 1.286 | 1 | 11 | 455 |
| GO:0070062 | extracellular vesicular exosome | 3.71e-01 | 1.00e+00 | 0.472 | 3 | 57 | 2400 |
| GO:0006351 | transcription, DNA-templated | 3.80e-01 | 1.00e+00 | 0.618 | 2 | 57 | 1446 |
| GO:0016032 | viral process | 3.88e-01 | 1.00e+00 | 1.055 | 1 | 26 | 534 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 4.08e-01 | 1.00e+00 | 0.961 | 1 | 37 | 570 |
| GO:0045087 | innate immune response | 4.22e-01 | 1.00e+00 | 0.896 | 1 | 15 | 596 |
| GO:0005730 | nucleolus | 4.45e-01 | 1.00e+00 | 0.435 | 2 | 66 | 1641 |
| GO:0007165 | signal transduction | 5.70e-01 | 1.00e+00 | 0.291 | 1 | 13 | 907 |
| GO:0005615 | extracellular space | 5.91e-01 | 1.00e+00 | 0.213 | 1 | 20 | 957 |
| GO:0008270 | zinc ion binding | 6.06e-01 | 1.00e+00 | 0.154 | 1 | 27 | 997 |
| GO:0046872 | metal ion binding | 7.09e-01 | 1.00e+00 | -0.236 | 1 | 29 | 1307 |
| GO:0005886 | plasma membrane | 9.23e-01 | 1.00e+00 | -1.219 | 1 | 49 | 2582 |