int-snw-11168
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.880 | 5.46e-16 | 2.81e-04 | 2.30e-03 |
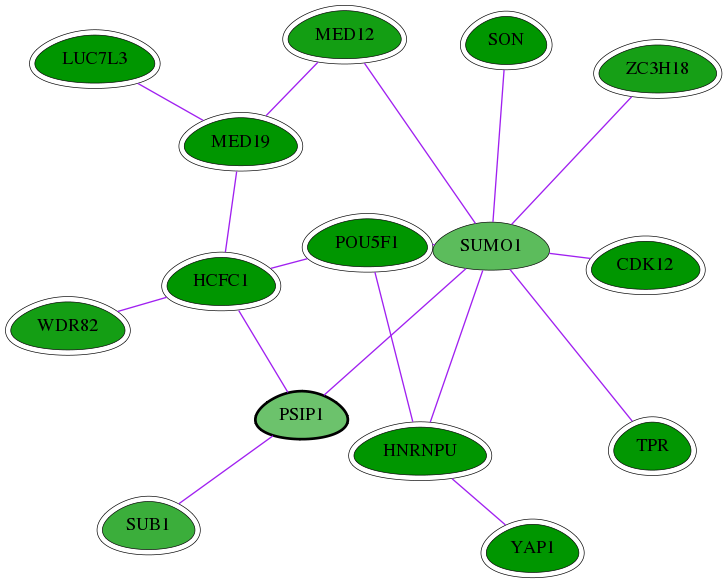
Genes (15)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| HCFC1 | 3054 | 40 | -2.925 | 3.449 | 71 | Yes | Yes |
| TPR | 7175 | 18 | -2.638 | 3.072 | 28 | Yes | Yes |
| SUB1 | 10923 | 2 | -2.042 | 2.880 | 27 | Yes | - |
| MED19 | 219541 | 30 | -2.657 | 3.449 | 115 | Yes | - |
| LUC7L3 | 51747 | 33 | -3.523 | 3.449 | 18 | Yes | - |
| ZC3H18 | 124245 | 3 | -2.426 | 2.937 | 6 | Yes | - |
| YAP1 | 10413 | 94 | -4.256 | 3.538 | 53 | Yes | - |
| MED12 | 9968 | 14 | -2.465 | 3.206 | 65 | Yes | - |
| WDR82 | 80335 | 7 | -2.455 | 3.129 | 64 | Yes | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| HNRNPU | 3192 | 76 | -2.869 | 3.538 | 139 | Yes | - |
| SON | 6651 | 9 | -3.844 | 3.108 | 9 | Yes | Yes |
| [ PSIP1 ] | 11168 | 1 | -1.537 | 2.880 | 17 | - | - |
| CDK12 | 51755 | 6 | -2.714 | 2.944 | 15 | Yes | - |
| SUMO1 | 7341 | 7 | -1.696 | 3.050 | 439 | - | - |
Interactions (17)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| HCFC1 | 3054 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| HCFC1 | 3054 | WDR82 | 80335 | pp | -- | int.I2D: BioGrid |
| SUMO1 | 7341 | MED12 | 9968 | pp | -- | int.I2D: BioGrid |
| HCFC1 | 3054 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| HNRNPU | 3192 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| SUMO1 | 7341 | PSIP1 | 11168 | pp | -- | int.Mint: MI:0914(association) |
| SON | 6651 | SUMO1 | 7341 | pp | -- | int.I2D: BioGrid |
| SUMO1 | 7341 | ZC3H18 | 124245 | pp | -- | int.I2D: BioGrid |
| SUMO1 | 7341 | CDK12 | 51755 | pp | -- | int.I2D: BioGrid |
| SUB1 | 10923 | PSIP1 | 11168 | pp | -- | int.I2D: BioGrid |
| HNRNPU | 3192 | SUMO1 | 7341 | pp | -- | int.I2D: BioGrid |
| HCFC1 | 3054 | PSIP1 | 11168 | pp | -- | int.I2D: HPRD, BCI; int.Ravasi: -; int.HPRD: in vitro |
| MED12 | 9968 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: BioGrid, IntAct |
| LUC7L3 | 51747 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| HNRNPU | 3192 | YAP1 | 10413 | pp | -- | int.I2D: HPRD; int.HPRD: in vitro, in vivo |
| POU5F1 | 5460 | SUMO1 | 7341 | pp | -- | int.I2D: BioGrid, BioGrid_Mouse |
| TPR | 7175 | SUMO1 | 7341 | pp | -- | int.I2D: BioGrid |
Related GO terms (224)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0044822 | poly(A) RNA binding | 1.96e-07 | 2.82e-03 | 3.035 | 9 | 50 | 1056 |
| GO:0005634 | nucleus | 1.04e-06 | 1.50e-02 | 1.562 | 14 | 159 | 4559 |
| GO:0005654 | nucleoplasm | 3.91e-06 | 5.63e-02 | 2.830 | 8 | 64 | 1082 |
| GO:0003682 | chromatin binding | 1.45e-05 | 2.09e-01 | 3.878 | 5 | 19 | 327 |
| GO:0016607 | nuclear speck | 2.41e-05 | 3.47e-01 | 4.483 | 4 | 12 | 172 |
| GO:0048188 | Set1C/COMPASS complex | 3.62e-05 | 5.22e-01 | 7.739 | 2 | 4 | 9 |
| GO:0034399 | nuclear periphery | 6.62e-05 | 9.55e-01 | 7.324 | 2 | 5 | 12 |
| GO:0008380 | RNA splicing | 7.24e-05 | 1.00e+00 | 4.076 | 4 | 22 | 228 |
| GO:0003713 | transcription coactivator activity | 8.41e-05 | 1.00e+00 | 4.020 | 4 | 24 | 237 |
| GO:0005730 | nucleolus | 8.49e-05 | 1.00e+00 | 2.229 | 8 | 66 | 1641 |
| GO:0001205 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 2.30e-04 | 1.00e+00 | 6.450 | 2 | 4 | 22 |
| GO:0005515 | protein binding | 4.61e-04 | 1.00e+00 | 1.053 | 13 | 198 | 6024 |
| GO:0001104 | RNA polymerase II transcription cofactor activity | 5.23e-04 | 1.00e+00 | 5.865 | 2 | 8 | 33 |
| GO:0016592 | mediator complex | 5.89e-04 | 1.00e+00 | 5.780 | 2 | 10 | 35 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 9.17e-04 | 1.00e+00 | 2.607 | 5 | 41 | 789 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:1901673 | regulation of spindle assembly involved in mitosis | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0043578 | nuclear matrix organization | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0010965 | regulation of mitotic sister chromatid separation | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0006404 | RNA import into nucleus | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0031453 | positive regulation of heterochromatin assembly | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0097100 | supercoiled DNA binding | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 1 |
| GO:0005643 | nuclear pore | 1.56e-03 | 1.00e+00 | 5.076 | 2 | 11 | 57 |
| GO:0000189 | MAPK import into nucleus | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0060242 | contact inhibition | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0031990 | mRNA export from nucleus in response to heat stress | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 2 | 2 |
| GO:0019046 | release from viral latency | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 2 | 2 |
| GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0090204 | protein localization to nuclear pore | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0070840 | dynein complex binding | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0002944 | cyclin K-CDK12 complex | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0046832 | negative regulation of RNA export from nucleus | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 2 |
| GO:0003729 | mRNA binding | 2.47e-03 | 1.00e+00 | 4.739 | 2 | 4 | 72 |
| GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 3 |
| GO:0006405 | RNA export from nucleus | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 2 | 3 |
| GO:0042306 | regulation of protein import into nucleus | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 2 | 3 |
| GO:0060913 | cardiac cell fate determination | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 3 |
| GO:0001650 | fibrillar center | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 3 |
| GO:0090245 | axis elongation involved in somitogenesis | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 3 |
| GO:0014044 | Schwann cell development | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 4 |
| GO:0000395 | mRNA 5'-splice site recognition | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 4 |
| GO:0030578 | PML body organization | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 4 |
| GO:0006999 | nuclear pore organization | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 4 |
| GO:0044615 | nuclear pore nuclear basket | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 4 |
| GO:0051726 | regulation of cell cycle | 4.44e-03 | 1.00e+00 | 4.309 | 2 | 5 | 97 |
| GO:0070461 | SAGA-type complex | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 2 | 5 |
| GO:0001714 | endodermal cell fate specification | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 1 | 5 |
| GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 2 | 5 |
| GO:0010793 | regulation of mRNA export from nucleus | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 1 | 5 |
| GO:0043405 | regulation of MAP kinase activity | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 1 | 5 |
| GO:0070849 | response to epidermal growth factor | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 1 | 5 |
| GO:0050733 | RS domain binding | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 3 | 6 |
| GO:0046825 | regulation of protein export from nucleus | 7.26e-03 | 1.00e+00 | 7.102 | 1 | 2 | 7 |
| GO:0072357 | PTW/PP1 phosphatase complex | 7.26e-03 | 1.00e+00 | 7.102 | 1 | 2 | 7 |
| GO:0051292 | nuclear pore complex assembly | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 4 | 8 |
| GO:0075713 | establishment of integrated proviral latency | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 8 |
| GO:0070688 | MLL5-L complex | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 8 |
| GO:0043254 | regulation of protein complex assembly | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 8 |
| GO:0035457 | cellular response to interferon-alpha | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 8 |
| GO:0006366 | transcription from RNA polymerase II promoter | 8.54e-03 | 1.00e+00 | 2.783 | 3 | 30 | 419 |
| GO:0043995 | histone acetyltransferase activity (H4-K5 specific) | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 5 | 9 |
| GO:0046972 | histone acetyltransferase activity (H4-K16 specific) | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 5 | 9 |
| GO:0042405 | nuclear inclusion body | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 2 | 9 |
| GO:0043495 | protein anchor | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 1 | 9 |
| GO:0043996 | histone acetyltransferase activity (H4-K8 specific) | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 5 | 9 |
| GO:0035413 | positive regulation of catenin import into nucleus | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 10 |
| GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 1 | 10 |
| GO:0019789 | SUMO ligase activity | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 1 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 4 | 11 |
| GO:0006397 | mRNA processing | 1.17e-02 | 1.00e+00 | 3.587 | 2 | 10 | 160 |
| GO:0045947 | negative regulation of translational initiation | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 2 | 12 |
| GO:0035198 | miRNA binding | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 12 |
| GO:0031965 | nuclear membrane | 1.28e-02 | 1.00e+00 | 3.517 | 2 | 10 | 168 |
| GO:0044212 | transcription regulatory region DNA binding | 1.28e-02 | 1.00e+00 | 3.517 | 2 | 17 | 168 |
| GO:0071480 | cellular response to gamma radiation | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 1 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 13 |
| GO:0051019 | mitogen-activated protein kinase binding | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 13 |
| GO:0031334 | positive regulation of protein complex assembly | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 13 |
| GO:0005868 | cytoplasmic dynein complex | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 1 | 13 |
| GO:0042800 | histone methyltransferase activity (H3-K4 specific) | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 3 | 13 |
| GO:0046827 | positive regulation of protein export from nucleus | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 1 | 13 |
| GO:0005667 | transcription factor complex | 1.39e-02 | 1.00e+00 | 3.458 | 2 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.43e-02 | 1.00e+00 | 3.433 | 2 | 5 | 178 |
| GO:0030518 | intracellular steroid hormone receptor signaling pathway | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 6 | 14 |
| GO:0005685 | U1 snRNP | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 14 |
| GO:0005487 | nucleocytoplasmic transporter activity | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 3 | 14 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.52e-02 | 1.00e+00 | 3.386 | 2 | 22 | 184 |
| GO:0043981 | histone H4-K5 acetylation | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 5 | 15 |
| GO:0042809 | vitamin D receptor binding | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 6 | 15 |
| GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 3 | 15 |
| GO:0042307 | positive regulation of protein import into nucleus | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 4 | 15 |
| GO:0001824 | blastocyst development | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 2 | 15 |
| GO:0043982 | histone H4-K8 acetylation | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 5 | 15 |
| GO:0006376 | mRNA splice site selection | 1.55e-02 | 1.00e+00 | 6.002 | 1 | 1 | 15 |
| GO:0060334 | regulation of interferon-gamma-mediated signaling pathway | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 16 |
| GO:0001190 | RNA polymerase II transcription factor binding transcription factor activity involved in positive regulation of transcription | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 2 | 16 |
| GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 16 |
| GO:0006355 | regulation of transcription, DNA-templated | 1.67e-02 | 1.00e+00 | 1.948 | 4 | 43 | 997 |
| GO:0051568 | histone H3-K4 methylation | 1.75e-02 | 1.00e+00 | 5.822 | 1 | 3 | 17 |
| GO:0090316 | positive regulation of intracellular protein transport | 1.75e-02 | 1.00e+00 | 5.822 | 1 | 2 | 17 |
| GO:0035327 | transcriptionally active chromatin | 1.75e-02 | 1.00e+00 | 5.822 | 1 | 2 | 17 |
| GO:0030332 | cyclin binding | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 1 | 18 |
| GO:0033613 | activating transcription factor binding | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 3 | 18 |
| GO:0043984 | histone H4-K16 acetylation | 1.96e-02 | 1.00e+00 | 5.661 | 1 | 5 | 19 |
| GO:0014003 | oligodendrocyte development | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 5 | 20 |
| GO:0019221 | cytokine-mediated signaling pathway | 2.15e-02 | 1.00e+00 | 3.121 | 2 | 8 | 221 |
| GO:0000123 | histone acetyltransferase complex | 2.16e-02 | 1.00e+00 | 5.517 | 1 | 6 | 21 |
| GO:0035097 | histone methyltransferase complex | 2.16e-02 | 1.00e+00 | 5.517 | 1 | 3 | 21 |
| GO:0045787 | positive regulation of cell cycle | 2.26e-02 | 1.00e+00 | 5.450 | 1 | 2 | 22 |
| GO:0043484 | regulation of RNA splicing | 2.26e-02 | 1.00e+00 | 5.450 | 1 | 1 | 22 |
| GO:0005720 | nuclear heterochromatin | 2.26e-02 | 1.00e+00 | 5.450 | 1 | 2 | 22 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 2.26e-02 | 1.00e+00 | 5.450 | 1 | 2 | 22 |
| GO:0001105 | RNA polymerase II transcription coactivator activity | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 2 | 24 |
| GO:0008134 | transcription factor binding | 2.55e-02 | 1.00e+00 | 2.990 | 2 | 18 | 242 |
| GO:0006611 | protein export from nucleus | 2.57e-02 | 1.00e+00 | 5.265 | 1 | 3 | 25 |
| GO:0016925 | protein sumoylation | 2.57e-02 | 1.00e+00 | 5.265 | 1 | 2 | 25 |
| GO:0043392 | negative regulation of DNA binding | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 4 | 26 |
| GO:0035329 | hippo signaling | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 5 | 26 |
| GO:0046966 | thyroid hormone receptor binding | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 6 | 26 |
| GO:0015631 | tubulin binding | 2.77e-02 | 1.00e+00 | 5.154 | 1 | 2 | 27 |
| GO:0071339 | MLL1 complex | 2.77e-02 | 1.00e+00 | 5.154 | 1 | 6 | 27 |
| GO:0034605 | cellular response to heat | 2.87e-02 | 1.00e+00 | 5.102 | 1 | 1 | 28 |
| GO:0072686 | mitotic spindle | 2.98e-02 | 1.00e+00 | 5.051 | 1 | 3 | 29 |
| GO:0010467 | gene expression | 2.98e-02 | 1.00e+00 | 2.108 | 3 | 45 | 669 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 3.00e-02 | 1.00e+00 | 2.865 | 2 | 23 | 264 |
| GO:0006606 | protein import into nucleus | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 2 | 30 |
| GO:0031647 | regulation of protein stability | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 4 | 30 |
| GO:0010827 | regulation of glucose transport | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 4 | 30 |
| GO:0004693 | cyclin-dependent protein serine/threonine kinase activity | 3.18e-02 | 1.00e+00 | 4.955 | 1 | 1 | 31 |
| GO:0007094 | mitotic spindle assembly checkpoint | 3.18e-02 | 1.00e+00 | 4.955 | 1 | 3 | 31 |
| GO:0003677 | DNA binding | 3.25e-02 | 1.00e+00 | 1.659 | 4 | 49 | 1218 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 3.28e-02 | 1.00e+00 | 4.909 | 1 | 2 | 32 |
| GO:0031072 | heat shock protein binding | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 2 | 33 |
| GO:0030178 | negative regulation of Wnt signaling pathway | 3.58e-02 | 1.00e+00 | 4.780 | 1 | 4 | 35 |
| GO:0007077 | mitotic nuclear envelope disassembly | 3.68e-02 | 1.00e+00 | 4.739 | 1 | 4 | 36 |
| GO:0032880 | regulation of protein localization | 3.78e-02 | 1.00e+00 | 4.700 | 1 | 2 | 37 |
| GO:0008645 | hexose transport | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 4 | 39 |
| GO:0009408 | response to heat | 4.08e-02 | 1.00e+00 | 4.587 | 1 | 6 | 40 |
| GO:0035019 | somatic stem cell maintenance | 4.08e-02 | 1.00e+00 | 4.587 | 1 | 6 | 40 |
| GO:0030521 | androgen receptor signaling pathway | 4.18e-02 | 1.00e+00 | 4.552 | 1 | 5 | 41 |
| GO:0019898 | extrinsic component of membrane | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 2 | 42 |
| GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity | 4.38e-02 | 1.00e+00 | 4.483 | 1 | 7 | 43 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 4.48e-02 | 1.00e+00 | 4.450 | 1 | 3 | 44 |
| GO:0003712 | transcription cofactor activity | 4.48e-02 | 1.00e+00 | 4.450 | 1 | 8 | 44 |
| GO:0019827 | stem cell maintenance | 4.78e-02 | 1.00e+00 | 4.355 | 1 | 7 | 47 |
| GO:0003723 | RNA binding | 4.80e-02 | 1.00e+00 | 2.491 | 2 | 18 | 342 |
| GO:0000910 | cytokinesis | 5.18e-02 | 1.00e+00 | 4.237 | 1 | 2 | 51 |
| GO:0000226 | microtubule cytoskeleton organization | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 2 | 52 |
| GO:0009611 | response to wounding | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 5 | 52 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 5.38e-02 | 1.00e+00 | 4.181 | 1 | 5 | 53 |
| GO:0015758 | glucose transport | 5.57e-02 | 1.00e+00 | 4.128 | 1 | 4 | 55 |
| GO:0006396 | RNA processing | 5.97e-02 | 1.00e+00 | 4.027 | 1 | 5 | 59 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 5.97e-02 | 1.00e+00 | 4.027 | 1 | 6 | 59 |
| GO:0008013 | beta-catenin binding | 6.06e-02 | 1.00e+00 | 4.002 | 1 | 9 | 60 |
| GO:0060333 | interferon-gamma-mediated signaling pathway | 6.26e-02 | 1.00e+00 | 3.955 | 1 | 3 | 62 |
| GO:0000776 | kinetochore | 6.36e-02 | 1.00e+00 | 3.932 | 1 | 5 | 63 |
| GO:0010468 | regulation of gene expression | 6.85e-02 | 1.00e+00 | 3.822 | 1 | 2 | 68 |
| GO:0003697 | single-stranded DNA binding | 6.85e-02 | 1.00e+00 | 3.822 | 1 | 5 | 68 |
| GO:0060021 | palate development | 7.24e-02 | 1.00e+00 | 3.739 | 1 | 1 | 72 |
| GO:0000785 | chromatin | 7.33e-02 | 1.00e+00 | 3.719 | 1 | 6 | 73 |
| GO:0001843 | neural tube closure | 7.43e-02 | 1.00e+00 | 3.700 | 1 | 2 | 74 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 7.43e-02 | 1.00e+00 | 3.700 | 1 | 4 | 74 |
| GO:0060070 | canonical Wnt signaling pathway | 7.53e-02 | 1.00e+00 | 3.680 | 1 | 4 | 75 |
| GO:0071013 | catalytic step 2 spliceosome | 7.82e-02 | 1.00e+00 | 3.624 | 1 | 7 | 78 |
| GO:0009653 | anatomical structure morphogenesis | 8.30e-02 | 1.00e+00 | 3.534 | 1 | 2 | 83 |
| GO:0006979 | response to oxidative stress | 8.68e-02 | 1.00e+00 | 3.466 | 1 | 6 | 87 |
| GO:0016605 | PML body | 8.78e-02 | 1.00e+00 | 3.450 | 1 | 3 | 88 |
| GO:0050821 | protein stabilization | 8.78e-02 | 1.00e+00 | 3.450 | 1 | 3 | 88 |
| GO:0001649 | osteoblast differentiation | 9.16e-02 | 1.00e+00 | 3.386 | 1 | 5 | 92 |
| GO:0016032 | viral process | 1.05e-01 | 1.00e+00 | 1.848 | 2 | 26 | 534 |
| GO:0003676 | nucleic acid binding | 1.06e-01 | 1.00e+00 | 3.168 | 1 | 2 | 107 |
| GO:0030529 | ribonucleoprotein complex | 1.10e-01 | 1.00e+00 | 3.102 | 1 | 5 | 112 |
| GO:0005635 | nuclear envelope | 1.11e-01 | 1.00e+00 | 3.089 | 1 | 5 | 113 |
| GO:0006325 | chromatin organization | 1.16e-01 | 1.00e+00 | 3.027 | 1 | 12 | 118 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 1.17e-01 | 1.00e+00 | 1.754 | 2 | 37 | 570 |
| GO:0005215 | transporter activity | 1.27e-01 | 1.00e+00 | 2.887 | 1 | 3 | 130 |
| GO:0045202 | synapse | 1.33e-01 | 1.00e+00 | 2.822 | 1 | 5 | 136 |
| GO:0007507 | heart development | 1.35e-01 | 1.00e+00 | 2.790 | 1 | 7 | 139 |
| GO:0010628 | positive regulation of gene expression | 1.42e-01 | 1.00e+00 | 2.719 | 1 | 7 | 146 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.46e-01 | 1.00e+00 | 2.671 | 1 | 7 | 151 |
| GO:0046777 | protein autophosphorylation | 1.52e-01 | 1.00e+00 | 2.605 | 1 | 4 | 158 |
| GO:0008022 | protein C-terminus binding | 1.55e-01 | 1.00e+00 | 2.578 | 1 | 8 | 161 |
| GO:0043687 | post-translational protein modification | 1.56e-01 | 1.00e+00 | 2.569 | 1 | 5 | 162 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.58e-01 | 1.00e+00 | 2.552 | 1 | 15 | 164 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 1.63e-01 | 1.00e+00 | 1.460 | 2 | 39 | 699 |
| GO:0007049 | cell cycle | 1.67e-01 | 1.00e+00 | 2.466 | 1 | 6 | 174 |
| GO:0004672 | protein kinase activity | 1.69e-01 | 1.00e+00 | 2.442 | 1 | 3 | 177 |
| GO:0003714 | transcription corepressor activity | 1.70e-01 | 1.00e+00 | 2.433 | 1 | 11 | 178 |
| GO:0019904 | protein domain specific binding | 1.71e-01 | 1.00e+00 | 2.425 | 1 | 8 | 179 |
| GO:0004872 | receptor activity | 1.80e-01 | 1.00e+00 | 2.347 | 1 | 8 | 189 |
| GO:0007067 | mitotic nuclear division | 2.12e-01 | 1.00e+00 | 2.083 | 1 | 6 | 227 |
| GO:0030425 | dendrite | 2.14e-01 | 1.00e+00 | 2.064 | 1 | 6 | 230 |
| GO:0043025 | neuronal cell body | 2.27e-01 | 1.00e+00 | 1.973 | 1 | 9 | 245 |
| GO:0005975 | carbohydrate metabolic process | 2.33e-01 | 1.00e+00 | 1.926 | 1 | 8 | 253 |
| GO:0006281 | DNA repair | 2.40e-01 | 1.00e+00 | 1.881 | 1 | 14 | 261 |
| GO:0016020 | membrane | 2.51e-01 | 1.00e+00 | 0.779 | 3 | 46 | 1681 |
| GO:0008283 | cell proliferation | 2.92e-01 | 1.00e+00 | 1.552 | 1 | 14 | 328 |
| GO:0043565 | sequence-specific DNA binding | 3.05e-01 | 1.00e+00 | 1.475 | 1 | 11 | 346 |
| GO:0005925 | focal adhesion | 3.20e-01 | 1.00e+00 | 1.393 | 1 | 9 | 366 |
| GO:0000278 | mitotic cell cycle | 3.38e-01 | 1.00e+00 | 1.298 | 1 | 15 | 391 |
| GO:0009986 | cell surface | 3.46e-01 | 1.00e+00 | 1.258 | 1 | 9 | 402 |
| GO:0005737 | cytoplasm | 3.51e-01 | 1.00e+00 | 0.352 | 5 | 124 | 3767 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.53e-01 | 1.00e+00 | 1.219 | 1 | 24 | 413 |
| GO:0043066 | negative regulation of apoptotic process | 3.61e-01 | 1.00e+00 | 1.181 | 1 | 16 | 424 |
| GO:0055085 | transmembrane transport | 3.68e-01 | 1.00e+00 | 1.144 | 1 | 8 | 435 |
| GO:0044267 | cellular protein metabolic process | 3.94e-01 | 1.00e+00 | 1.020 | 1 | 20 | 474 |
| GO:0005524 | ATP binding | 3.97e-01 | 1.00e+00 | 0.567 | 2 | 31 | 1298 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 3.97e-01 | 1.00e+00 | 1.008 | 1 | 24 | 478 |
| GO:0042802 | identical protein binding | 4.01e-01 | 1.00e+00 | 0.990 | 1 | 19 | 484 |
| GO:0006351 | transcription, DNA-templated | 4.52e-01 | 1.00e+00 | 0.411 | 2 | 57 | 1446 |
| GO:0042803 | protein homodimerization activity | 4.69e-01 | 1.00e+00 | 0.692 | 1 | 19 | 595 |
| GO:0005829 | cytosol | 4.95e-01 | 1.00e+00 | 0.209 | 3 | 86 | 2496 |
| GO:0005739 | mitochondrion | 6.59e-01 | 1.00e+00 | -0.054 | 1 | 23 | 998 |
| GO:0044281 | small molecule metabolic process | 7.32e-01 | 1.00e+00 | -0.333 | 1 | 20 | 1211 |
| GO:0046872 | metal ion binding | 7.60e-01 | 1.00e+00 | -0.443 | 1 | 29 | 1307 |
| GO:0070062 | extracellular vesicular exosome | 9.35e-01 | 1.00e+00 | -1.320 | 1 | 57 | 2400 |
| GO:0005886 | plasma membrane | 9.48e-01 | 1.00e+00 | -1.425 | 1 | 49 | 2582 |