int-snw-10379
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.844 | 1.32e-15 | 4.34e-04 | 3.13e-03 |
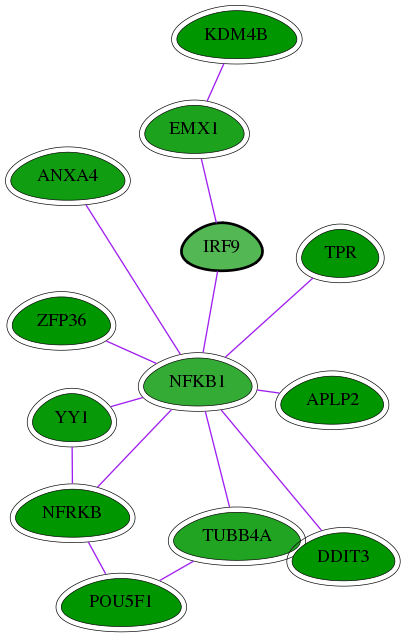
Genes (13)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| TUBB4A | 10382 | 7 | -2.312 | 2.975 | 88 | Yes | - |
| TPR | 7175 | 18 | -2.638 | 3.072 | 28 | Yes | Yes |
| APLP2 | 334 | 12 | -2.732 | 3.206 | 35 | Yes | Yes |
| EMX1 | 2016 | 4 | -2.369 | 3.216 | 19 | Yes | - |
| KDM4B | 23030 | 5 | -3.892 | 3.216 | 5 | Yes | - |
| ANXA4 | 307 | 7 | -2.478 | 3.062 | 15 | Yes | Yes |
| ZFP36 | 7538 | 10 | -2.968 | 2.869 | 24 | Yes | Yes |
| NFKB1 | 4790 | 9 | -2.123 | 2.870 | 263 | Yes | - |
| [ IRF9 ] | 10379 | 1 | -1.789 | 2.844 | 32 | - | - |
| NFRKB | 4798 | 48 | -3.129 | 3.108 | 23 | Yes | Yes |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| YY1 | 7528 | 24 | -2.572 | 3.120 | 114 | Yes | Yes |
| DDIT3 | 1649 | 60 | -2.825 | 3.504 | 343 | Yes | Yes |
Interactions (15)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| NFRKB | 4798 | YY1 | 7528 | pp | -- | int.I2D: BioGrid |
| NFKB1 | 4790 | IRF9 | 10379 | pp | -- | int.I2D: BioGrid |
| EMX1 | 2016 | KDM4B | 23030 | pp | -- | int.I2D: BioGrid_Mouse |
| NFKB1 | 4790 | ZFP36 | 7538 | pp | -- | int.I2D: BioGrid |
| DDIT3 | 1649 | NFKB1 | 4790 | pp | -- | int.I2D: BEHRENDS_AUTOPHAGY_LOW |
| APLP2 | 334 | NFKB1 | 4790 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| POU5F1 | 5460 | TUBB4A | 10382 | pp | -- | int.I2D: BioGrid |
| DDIT3 | 1649 | TUBB4A | 10382 | pp | -- | int.I2D: BEHRENDS_AUTOPHAGY_LOW |
| NFKB1 | 4790 | TPR | 7175 | pp | -- | int.I2D: MINT; int.Mint: MI:0915(physical association) |
| NFRKB | 4798 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| NFKB1 | 4790 | YY1 | 7528 | pp | -- | int.Intact: MI:0915(physical association) |
| NFKB1 | 4790 | TUBB4A | 10382 | pp | -- | int.I2D: MINT; int.Mint: MI:0915(physical association) |
| EMX1 | 2016 | IRF9 | 10379 | pp | -- | int.I2D: BioGrid_Mouse |
| ANXA4 | 307 | NFKB1 | 4790 | pp | -- | int.I2D: BioGrid, INNATEDB |
| NFKB1 | 4790 | NFRKB | 4798 | pp | -- | int.I2D: BCI, HPRD; int.Ravasi: -; int.HPRD: in vitro |
Related GO terms (278)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0005634 | nucleus | 3.11e-07 | 4.48e-03 | 1.662 | 13 | 159 | 4559 |
| GO:0044212 | transcription regulatory region DNA binding | 1.17e-05 | 1.69e-01 | 4.723 | 4 | 17 | 168 |
| GO:0000975 | regulatory region DNA binding | 2.69e-05 | 3.88e-01 | 7.946 | 2 | 2 | 9 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 5.82e-05 | 8.39e-01 | 7.415 | 2 | 2 | 13 |
| GO:0031011 | Ino80 complex | 6.78e-05 | 9.78e-01 | 7.308 | 2 | 6 | 14 |
| GO:0006355 | regulation of transcription, DNA-templated | 1.21e-04 | 1.00e+00 | 2.739 | 6 | 43 | 997 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 2.45e-04 | 1.00e+00 | 2.988 | 5 | 39 | 699 |
| GO:0042594 | response to starvation | 3.44e-04 | 1.00e+00 | 6.161 | 2 | 4 | 31 |
| GO:0003677 | DNA binding | 3.66e-04 | 1.00e+00 | 2.450 | 6 | 49 | 1218 |
| GO:0031072 | heat shock protein binding | 3.90e-04 | 1.00e+00 | 6.071 | 2 | 2 | 33 |
| GO:0006366 | transcription from RNA polymerase II promoter | 4.08e-04 | 1.00e+00 | 3.405 | 4 | 30 | 419 |
| GO:1901673 | regulation of spindle assembly involved in mitosis | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0043578 | nuclear matrix organization | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:2000016 | negative regulation of determination of dorsal identity | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0010965 | regulation of mitotic sister chromatid separation | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0045083 | negative regulation of interleukin-12 biosynthetic process | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0006404 | RNA import into nucleus | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0031453 | positive regulation of heterochromatin assembly | 9.01e-04 | 1.00e+00 | 10.116 | 1 | 1 | 1 |
| GO:0050728 | negative regulation of inflammatory response | 1.12e-03 | 1.00e+00 | 5.308 | 2 | 3 | 56 |
| GO:0008134 | transcription factor binding | 1.18e-03 | 1.00e+00 | 3.782 | 3 | 18 | 242 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 1.30e-03 | 1.00e+00 | 2.961 | 4 | 37 | 570 |
| GO:0031990 | mRNA export from nucleus in response to heat stress | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 2 | 2 |
| GO:0006403 | RNA localization | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:2000483 | negative regulation of interleukin-8 secretion | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0070840 | dynein complex binding | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0046832 | negative regulation of RNA export from nucleus | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0034696 | response to prostaglandin F | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0000189 | MAPK import into nucleus | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0044324 | regulation of transcription involved in anterior/posterior axis specification | 1.80e-03 | 1.00e+00 | 9.116 | 1 | 1 | 2 |
| GO:0005654 | nucleoplasm | 1.82e-03 | 1.00e+00 | 2.358 | 5 | 64 | 1082 |
| GO:0003729 | mRNA binding | 1.85e-03 | 1.00e+00 | 4.946 | 2 | 4 | 72 |
| GO:0005515 | protein binding | 2.00e-03 | 1.00e+00 | 1.019 | 11 | 198 | 6024 |
| GO:0046914 | transition metal ion binding | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0042306 | regulation of protein import into nucleus | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 2 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:1900127 | positive regulation of hyaluronan biosynthetic process | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0050779 | RNA destabilization | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0033256 | I-kappaB/NF-kappaB complex | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0006405 | RNA export from nucleus | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 2 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.70e-03 | 1.00e+00 | 8.531 | 1 | 1 | 3 |
| GO:0043565 | sequence-specific DNA binding | 3.27e-03 | 1.00e+00 | 3.266 | 3 | 11 | 346 |
| GO:0032680 | regulation of tumor necrosis factor production | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0032375 | negative regulation of cholesterol transport | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0010957 | negative regulation of vitamin D biosynthetic process | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0032269 | negative regulation of cellular protein metabolic process | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:2000630 | positive regulation of miRNA metabolic process | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0006999 | nuclear pore organization | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0033269 | internode region of axon | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0044615 | nuclear pore nuclear basket | 3.60e-03 | 1.00e+00 | 8.116 | 1 | 1 | 4 |
| GO:0001714 | endodermal cell fate specification | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0021796 | cerebral cortex regionalization | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:1990138 | neuron projection extension | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0019957 | C-C chemokine binding | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0043620 | regulation of DNA-templated transcription in response to stress | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0010884 | positive regulation of lipid storage | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0035925 | mRNA 3'-UTR AU-rich region binding | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0045351 | type I interferon biosynthetic process | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0010793 | regulation of mRNA export from nucleus | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0070849 | response to epidermal growth factor | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 5 |
| GO:0004859 | phospholipase inhibitor activity | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 1 | 6 |
| GO:0046825 | regulation of protein export from nucleus | 6.29e-03 | 1.00e+00 | 7.308 | 1 | 2 | 7 |
| GO:0071354 | cellular response to interleukin-6 | 6.29e-03 | 1.00e+00 | 7.308 | 1 | 2 | 7 |
| GO:0051292 | nuclear pore complex assembly | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 4 | 8 |
| GO:0007617 | mating behavior | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0001955 | blood vessel maturation | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0071316 | cellular response to nicotine | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0035457 | cellular response to interferon-alpha | 7.19e-03 | 1.00e+00 | 7.116 | 1 | 1 | 8 |
| GO:0006974 | cellular response to DNA damage stimulus | 7.87e-03 | 1.00e+00 | 3.877 | 2 | 7 | 151 |
| GO:0042405 | nuclear inclusion body | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 2 | 9 |
| GO:0010225 | response to UV-C | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 2 | 9 |
| GO:0035278 | negative regulation of translation involved in gene silencing by miRNA | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0006983 | ER overload response | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0043495 | protein anchor | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0000400 | four-way junction DNA binding | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:0021895 | cerebral cortex neuron differentiation | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 1 | 9 |
| GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 10 |
| GO:0035413 | positive regulation of catenin import into nucleus | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 10 |
| GO:0031965 | nuclear membrane | 9.67e-03 | 1.00e+00 | 3.723 | 2 | 10 | 168 |
| GO:0045662 | negative regulation of myoblast differentiation | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 3 | 11 |
| GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 2 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 4 | 11 |
| GO:0005667 | transcription factor complex | 1.05e-02 | 1.00e+00 | 3.664 | 2 | 17 | 175 |
| GO:0006878 | cellular copper ion homeostasis | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 12 |
| GO:0006402 | mRNA catabolic process | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 12 |
| GO:0045947 | negative regulation of translational initiation | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 2 | 12 |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 3 | 12 |
| GO:0003714 | transcription corepressor activity | 1.08e-02 | 1.00e+00 | 3.640 | 2 | 11 | 178 |
| GO:0034399 | nuclear periphery | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 5 | 12 |
| GO:0035198 | miRNA binding | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 12 |
| GO:0046688 | response to copper ion | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0051213 | dioxygenase activity | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0051019 | mitogen-activated protein kinase binding | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 2 | 13 |
| GO:0005868 | cytoplasmic dynein complex | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0046827 | positive regulation of protein export from nucleus | 1.17e-02 | 1.00e+00 | 6.415 | 1 | 1 | 13 |
| GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 14 |
| GO:0001967 | suckling behavior | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 14 |
| GO:0031293 | membrane protein intracellular domain proteolysis | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 14 |
| GO:0017091 | AU-rich element binding | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 2 | 14 |
| GO:0071375 | cellular response to peptide hormone stimulus | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 2 | 14 |
| GO:0005487 | nucleocytoplasmic transporter activity | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 3 | 14 |
| GO:0042307 | positive regulation of protein import into nucleus | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 4 | 15 |
| GO:0001824 | blastocyst development | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 15 |
| GO:0048854 | brain morphogenesis | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 15 |
| GO:0043209 | myelin sheath | 1.43e-02 | 1.00e+00 | 6.116 | 1 | 1 | 16 |
| GO:0051276 | chromosome organization | 1.43e-02 | 1.00e+00 | 6.116 | 1 | 2 | 16 |
| GO:0045638 | negative regulation of myeloid cell differentiation | 1.43e-02 | 1.00e+00 | 6.116 | 1 | 1 | 16 |
| GO:0048593 | camera-type eye morphogenesis | 1.43e-02 | 1.00e+00 | 6.116 | 1 | 1 | 16 |
| GO:0005829 | cytosol | 1.51e-02 | 1.00e+00 | 1.415 | 6 | 86 | 2496 |
| GO:0012506 | vesicle membrane | 1.52e-02 | 1.00e+00 | 6.028 | 1 | 1 | 17 |
| GO:0090316 | positive regulation of intracellular protein transport | 1.52e-02 | 1.00e+00 | 6.028 | 1 | 2 | 17 |
| GO:0071889 | 14-3-3 protein binding | 1.61e-02 | 1.00e+00 | 5.946 | 1 | 2 | 18 |
| GO:0019221 | cytokine-mediated signaling pathway | 1.63e-02 | 1.00e+00 | 3.328 | 2 | 8 | 221 |
| GO:0032757 | positive regulation of interleukin-8 production | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:0043393 | regulation of protein binding | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 20 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2.05e-02 | 1.00e+00 | 5.592 | 1 | 1 | 23 |
| GO:0006611 | protein export from nucleus | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 3 | 25 |
| GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 3 | 25 |
| GO:0031519 | PcG protein complex | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 1 | 25 |
| GO:0051059 | NF-kappaB binding | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 3 | 25 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 2.28e-02 | 1.00e+00 | 3.071 | 2 | 23 | 264 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 2.32e-02 | 1.00e+00 | 5.415 | 1 | 3 | 26 |
| GO:0051209 | release of sequestered calcium ion into cytosol | 2.32e-02 | 1.00e+00 | 5.415 | 1 | 1 | 26 |
| GO:0015631 | tubulin binding | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 2 | 27 |
| GO:0007017 | microtubule-based process | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 2 | 27 |
| GO:0051258 | protein polymerization | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 2 | 27 |
| GO:0006954 | inflammatory response | 2.49e-02 | 1.00e+00 | 3.002 | 2 | 6 | 277 |
| GO:0003727 | single-stranded RNA binding | 2.50e-02 | 1.00e+00 | 5.308 | 1 | 2 | 28 |
| GO:0034605 | cellular response to heat | 2.50e-02 | 1.00e+00 | 5.308 | 1 | 1 | 28 |
| GO:0030901 | midbrain development | 2.58e-02 | 1.00e+00 | 5.258 | 1 | 1 | 29 |
| GO:0072686 | mitotic spindle | 2.58e-02 | 1.00e+00 | 5.258 | 1 | 3 | 29 |
| GO:0006606 | protein import into nucleus | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 2 | 30 |
| GO:0031647 | regulation of protein stability | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 4 | 30 |
| GO:0010827 | regulation of glucose transport | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 4 | 30 |
| GO:0034644 | cellular response to UV | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 1 | 30 |
| GO:0005544 | calcium-dependent phospholipid binding | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 1 | 30 |
| GO:0010494 | cytoplasmic stress granule | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 2 | 30 |
| GO:0007094 | mitotic spindle assembly checkpoint | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 3 | 31 |
| GO:0071347 | cellular response to interleukin-1 | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 2 | 31 |
| GO:0005737 | cytoplasm | 3.06e-02 | 1.00e+00 | 1.044 | 7 | 124 | 3767 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 3.09e-02 | 1.00e+00 | 2.077 | 3 | 41 | 789 |
| GO:0007077 | mitotic nuclear envelope disassembly | 3.20e-02 | 1.00e+00 | 4.946 | 1 | 4 | 36 |
| GO:0051084 | 'de novo' posttranslational protein folding | 3.29e-02 | 1.00e+00 | 4.906 | 1 | 5 | 37 |
| GO:0008645 | hexose transport | 3.46e-02 | 1.00e+00 | 4.830 | 1 | 4 | 39 |
| GO:0035019 | somatic stem cell maintenance | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 6 | 40 |
| GO:0050885 | neuromuscular process controlling balance | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 2 | 40 |
| GO:0019898 | extrinsic component of membrane | 3.72e-02 | 1.00e+00 | 4.723 | 1 | 2 | 42 |
| GO:0043525 | positive regulation of neuron apoptotic process | 4.07e-02 | 1.00e+00 | 4.592 | 1 | 4 | 46 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 4.24e-02 | 1.00e+00 | 4.531 | 1 | 3 | 48 |
| GO:0030900 | forebrain development | 4.33e-02 | 1.00e+00 | 4.501 | 1 | 1 | 49 |
| GO:0006986 | response to unfolded protein | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 4 | 50 |
| GO:0034976 | response to endoplasmic reticulum stress | 4.50e-02 | 1.00e+00 | 4.443 | 1 | 2 | 51 |
| GO:0045454 | cell redox homeostasis | 4.50e-02 | 1.00e+00 | 4.443 | 1 | 1 | 51 |
| GO:0009611 | response to wounding | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 5 | 52 |
| GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4.68e-02 | 1.00e+00 | 4.388 | 1 | 4 | 53 |
| GO:0000278 | mitotic cell cycle | 4.69e-02 | 1.00e+00 | 2.505 | 2 | 15 | 391 |
| GO:0015758 | glucose transport | 4.85e-02 | 1.00e+00 | 4.334 | 1 | 4 | 55 |
| GO:0000724 | double-strand break repair via homologous recombination | 4.85e-02 | 1.00e+00 | 4.334 | 1 | 2 | 55 |
| GO:0048306 | calcium-dependent protein binding | 4.93e-02 | 1.00e+00 | 4.308 | 1 | 2 | 56 |
| GO:0008203 | cholesterol metabolic process | 5.02e-02 | 1.00e+00 | 4.283 | 1 | 2 | 57 |
| GO:0005643 | nuclear pore | 5.02e-02 | 1.00e+00 | 4.283 | 1 | 11 | 57 |
| GO:0060337 | type I interferon signaling pathway | 5.11e-02 | 1.00e+00 | 4.258 | 1 | 2 | 58 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 5.19e-02 | 1.00e+00 | 4.233 | 1 | 6 | 59 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 5.19e-02 | 1.00e+00 | 4.233 | 1 | 5 | 59 |
| GO:0030855 | epithelial cell differentiation | 5.36e-02 | 1.00e+00 | 4.185 | 1 | 1 | 61 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 5.36e-02 | 1.00e+00 | 4.185 | 1 | 2 | 61 |
| GO:0032481 | positive regulation of type I interferon production | 5.36e-02 | 1.00e+00 | 4.185 | 1 | 6 | 61 |
| GO:0043066 | negative regulation of apoptotic process | 5.43e-02 | 1.00e+00 | 2.388 | 2 | 16 | 424 |
| GO:0060333 | interferon-gamma-mediated signaling pathway | 5.45e-02 | 1.00e+00 | 4.161 | 1 | 3 | 62 |
| GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 5.53e-02 | 1.00e+00 | 4.138 | 1 | 3 | 63 |
| GO:0004867 | serine-type endopeptidase inhibitor activity | 5.53e-02 | 1.00e+00 | 4.138 | 1 | 3 | 63 |
| GO:0000776 | kinetochore | 5.53e-02 | 1.00e+00 | 4.138 | 1 | 5 | 63 |
| GO:0006310 | DNA recombination | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 7 | 65 |
| GO:0071260 | cellular response to mechanical stimulus | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 4 | 65 |
| GO:0034166 | toll-like receptor 10 signaling pathway | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 4 | 65 |
| GO:0034146 | toll-like receptor 5 signaling pathway | 5.71e-02 | 1.00e+00 | 4.093 | 1 | 4 | 65 |
| GO:0010468 | regulation of gene expression | 5.96e-02 | 1.00e+00 | 4.028 | 1 | 2 | 68 |
| GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway | 6.22e-02 | 1.00e+00 | 3.966 | 1 | 4 | 71 |
| GO:0007626 | locomotory behavior | 6.22e-02 | 1.00e+00 | 3.966 | 1 | 2 | 71 |
| GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway | 6.22e-02 | 1.00e+00 | 3.966 | 1 | 4 | 71 |
| GO:0043086 | negative regulation of catalytic activity | 6.30e-02 | 1.00e+00 | 3.946 | 1 | 2 | 72 |
| GO:0034162 | toll-like receptor 9 signaling pathway | 6.30e-02 | 1.00e+00 | 3.946 | 1 | 4 | 72 |
| GO:0009791 | post-embryonic development | 6.39e-02 | 1.00e+00 | 3.926 | 1 | 3 | 73 |
| GO:0034134 | toll-like receptor 2 signaling pathway | 6.39e-02 | 1.00e+00 | 3.926 | 1 | 4 | 73 |
| GO:0044822 | poly(A) RNA binding | 6.44e-02 | 1.00e+00 | 1.656 | 3 | 50 | 1056 |
| GO:0002020 | protease binding | 6.47e-02 | 1.00e+00 | 3.906 | 1 | 6 | 74 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 6.47e-02 | 1.00e+00 | 3.906 | 1 | 4 | 74 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 6.56e-02 | 1.00e+00 | 3.887 | 1 | 1 | 75 |
| GO:0044267 | cellular protein metabolic process | 6.62e-02 | 1.00e+00 | 2.227 | 2 | 20 | 474 |
| GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 6.64e-02 | 1.00e+00 | 3.868 | 1 | 3 | 76 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 6.72e-02 | 1.00e+00 | 2.215 | 2 | 24 | 478 |
| GO:0005929 | cilium | 6.72e-02 | 1.00e+00 | 3.849 | 1 | 2 | 77 |
| GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 6.81e-02 | 1.00e+00 | 3.830 | 1 | 3 | 78 |
| GO:0042802 | identical protein binding | 6.87e-02 | 1.00e+00 | 2.197 | 2 | 19 | 484 |
| GO:0034138 | toll-like receptor 3 signaling pathway | 6.89e-02 | 1.00e+00 | 3.812 | 1 | 3 | 79 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 6.89e-02 | 1.00e+00 | 3.812 | 1 | 2 | 79 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 6.98e-02 | 1.00e+00 | 3.794 | 1 | 5 | 80 |
| GO:0071222 | cellular response to lipopolysaccharide | 7.06e-02 | 1.00e+00 | 3.776 | 1 | 4 | 81 |
| GO:0009653 | anatomical structure morphogenesis | 7.23e-02 | 1.00e+00 | 3.741 | 1 | 2 | 83 |
| GO:0009952 | anterior/posterior pattern specification | 7.23e-02 | 1.00e+00 | 3.741 | 1 | 4 | 83 |
| GO:0050852 | T cell receptor signaling pathway | 7.40e-02 | 1.00e+00 | 3.706 | 1 | 2 | 85 |
| GO:0003690 | double-stranded DNA binding | 7.48e-02 | 1.00e+00 | 3.689 | 1 | 3 | 86 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 7.48e-02 | 1.00e+00 | 3.689 | 1 | 8 | 86 |
| GO:0006979 | response to oxidative stress | 7.57e-02 | 1.00e+00 | 3.673 | 1 | 6 | 87 |
| GO:0016363 | nuclear matrix | 7.90e-02 | 1.00e+00 | 3.608 | 1 | 6 | 91 |
| GO:0005200 | structural constituent of cytoskeleton | 7.90e-02 | 1.00e+00 | 3.608 | 1 | 6 | 91 |
| GO:0005770 | late endosome | 8.07e-02 | 1.00e+00 | 3.576 | 1 | 2 | 93 |
| GO:0034142 | toll-like receptor 4 signaling pathway | 8.32e-02 | 1.00e+00 | 3.531 | 1 | 4 | 96 |
| GO:0016568 | chromatin modification | 8.49e-02 | 1.00e+00 | 3.501 | 1 | 4 | 98 |
| GO:0010951 | negative regulation of endopeptidase activity | 8.98e-02 | 1.00e+00 | 3.415 | 1 | 6 | 104 |
| GO:0002224 | toll-like receptor signaling pathway | 9.39e-02 | 1.00e+00 | 3.347 | 1 | 4 | 109 |
| GO:0005635 | nuclear envelope | 9.72e-02 | 1.00e+00 | 3.295 | 1 | 5 | 113 |
| GO:0042803 | protein homodimerization activity | 9.82e-02 | 1.00e+00 | 1.899 | 2 | 19 | 595 |
| GO:0008201 | heparin binding | 1.05e-01 | 1.00e+00 | 3.185 | 1 | 1 | 122 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 1.06e-01 | 1.00e+00 | 3.161 | 1 | 5 | 124 |
| GO:0007050 | cell cycle arrest | 1.06e-01 | 1.00e+00 | 3.161 | 1 | 5 | 124 |
| GO:0051607 | defense response to virus | 1.08e-01 | 1.00e+00 | 3.138 | 1 | 1 | 126 |
| GO:0005215 | transporter activity | 1.11e-01 | 1.00e+00 | 3.093 | 1 | 3 | 130 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 1.16e-01 | 1.00e+00 | 3.028 | 1 | 4 | 136 |
| GO:0016055 | Wnt signaling pathway | 1.18e-01 | 1.00e+00 | 3.007 | 1 | 4 | 138 |
| GO:0006457 | protein folding | 1.22e-01 | 1.00e+00 | 2.956 | 1 | 7 | 143 |
| GO:0007166 | cell surface receptor signaling pathway | 1.25e-01 | 1.00e+00 | 2.916 | 1 | 4 | 147 |
| GO:0038095 | Fc-epsilon receptor signaling pathway | 1.41e-01 | 1.00e+00 | 2.723 | 1 | 5 | 168 |
| GO:0031625 | ubiquitin protein ligase binding | 1.49e-01 | 1.00e+00 | 2.640 | 1 | 5 | 178 |
| GO:0003924 | GTPase activity | 1.64e-01 | 1.00e+00 | 2.494 | 1 | 7 | 197 |
| GO:0001701 | in utero embryonic development | 1.67e-01 | 1.00e+00 | 2.465 | 1 | 8 | 201 |
| GO:0006184 | GTP catabolic process | 1.77e-01 | 1.00e+00 | 2.374 | 1 | 7 | 214 |
| GO:0016071 | mRNA metabolic process | 1.83e-01 | 1.00e+00 | 2.315 | 1 | 8 | 223 |
| GO:0007067 | mitotic nuclear division | 1.86e-01 | 1.00e+00 | 2.289 | 1 | 6 | 227 |
| GO:0003713 | transcription coactivator activity | 1.94e-01 | 1.00e+00 | 2.227 | 1 | 24 | 237 |
| GO:0043025 | neuronal cell body | 2.00e-01 | 1.00e+00 | 2.179 | 1 | 9 | 245 |
| GO:0016070 | RNA metabolic process | 2.01e-01 | 1.00e+00 | 2.167 | 1 | 8 | 247 |
| GO:0005874 | microtubule | 2.03e-01 | 1.00e+00 | 2.156 | 1 | 7 | 249 |
| GO:0005975 | carbohydrate metabolic process | 2.06e-01 | 1.00e+00 | 2.133 | 1 | 8 | 253 |
| GO:0006281 | DNA repair | 2.11e-01 | 1.00e+00 | 2.088 | 1 | 14 | 261 |
| GO:0007283 | spermatogenesis | 2.12e-01 | 1.00e+00 | 2.082 | 1 | 3 | 262 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 2.17e-01 | 1.00e+00 | 2.044 | 1 | 7 | 269 |
| GO:0008270 | zinc ion binding | 2.25e-01 | 1.00e+00 | 1.154 | 2 | 27 | 997 |
| GO:0030198 | extracellular matrix organization | 2.27e-01 | 1.00e+00 | 1.971 | 1 | 6 | 283 |
| GO:0042493 | response to drug | 2.28e-01 | 1.00e+00 | 1.966 | 1 | 6 | 284 |
| GO:0007186 | G-protein coupled receptor signaling pathway | 2.31e-01 | 1.00e+00 | 1.941 | 1 | 1 | 289 |
| GO:0030154 | cell differentiation | 2.40e-01 | 1.00e+00 | 1.882 | 1 | 5 | 301 |
| GO:0035556 | intracellular signal transduction | 2.41e-01 | 1.00e+00 | 1.872 | 1 | 9 | 303 |
| GO:0005525 | GTP binding | 2.46e-01 | 1.00e+00 | 1.840 | 1 | 12 | 310 |
| GO:0019901 | protein kinase binding | 2.51e-01 | 1.00e+00 | 1.807 | 1 | 18 | 317 |
| GO:0003682 | chromatin binding | 2.58e-01 | 1.00e+00 | 1.762 | 1 | 19 | 327 |
| GO:0030054 | cell junction | 2.64e-01 | 1.00e+00 | 1.723 | 1 | 5 | 336 |
| GO:0003723 | RNA binding | 2.68e-01 | 1.00e+00 | 1.698 | 1 | 18 | 342 |
| GO:0005925 | focal adhesion | 2.84e-01 | 1.00e+00 | 1.600 | 1 | 9 | 366 |
| GO:0046982 | protein heterodimerization activity | 2.93e-01 | 1.00e+00 | 1.546 | 1 | 8 | 380 |
| GO:0009986 | cell surface | 3.08e-01 | 1.00e+00 | 1.465 | 1 | 9 | 402 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.15e-01 | 1.00e+00 | 1.426 | 1 | 24 | 413 |
| GO:0055114 | oxidation-reduction process | 3.19e-01 | 1.00e+00 | 1.401 | 1 | 10 | 420 |
| GO:0055085 | transmembrane transport | 3.29e-01 | 1.00e+00 | 1.351 | 1 | 8 | 435 |
| GO:0048471 | perinuclear region of cytoplasm | 3.69e-01 | 1.00e+00 | 1.144 | 1 | 8 | 502 |
| GO:0070062 | extracellular vesicular exosome | 3.71e-01 | 1.00e+00 | 0.472 | 3 | 57 | 2400 |
| GO:0006351 | transcription, DNA-templated | 3.80e-01 | 1.00e+00 | 0.618 | 2 | 57 | 1446 |
| GO:0005509 | calcium ion binding | 3.86e-01 | 1.00e+00 | 1.063 | 1 | 11 | 531 |
| GO:0016032 | viral process | 3.88e-01 | 1.00e+00 | 1.055 | 1 | 26 | 534 |
| GO:0006915 | apoptotic process | 4.00e-01 | 1.00e+00 | 0.999 | 1 | 12 | 555 |
| GO:0045087 | innate immune response | 4.22e-01 | 1.00e+00 | 0.896 | 1 | 15 | 596 |
| GO:0005730 | nucleolus | 4.45e-01 | 1.00e+00 | 0.435 | 2 | 66 | 1641 |
| GO:0010467 | gene expression | 4.61e-01 | 1.00e+00 | 0.730 | 1 | 45 | 669 |
| GO:0007165 | signal transduction | 5.70e-01 | 1.00e+00 | 0.291 | 1 | 13 | 907 |
| GO:0005739 | mitochondrion | 6.06e-01 | 1.00e+00 | 0.153 | 1 | 23 | 998 |
| GO:0044281 | small molecule metabolic process | 6.80e-01 | 1.00e+00 | -0.126 | 1 | 20 | 1211 |
| GO:0005886 | plasma membrane | 7.05e-01 | 1.00e+00 | -0.219 | 2 | 49 | 2582 |
| GO:0046872 | metal ion binding | 7.09e-01 | 1.00e+00 | -0.236 | 1 | 29 | 1307 |
| GO:0016020 | membrane | 8.00e-01 | 1.00e+00 | -0.599 | 1 | 46 | 1681 |
| GO:0016021 | integral component of membrane | 8.54e-01 | 1.00e+00 | -0.837 | 1 | 17 | 1982 |