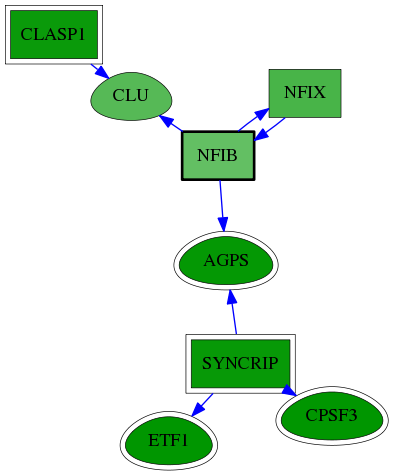
| GO:0003723 | RNA binding | 8.15e-04 | 1.00e+00 | 3.890 | 3 | 22 | 247 |
| GO:0016149 | translation release factor activity, codon specific | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0031592 | centrosomal corona | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0061518 | microglial cell proliferation | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:1902949 | positive regulation of tau-protein kinase activity | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:1902847 | regulation of neuronal signal transduction | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0021740 | principal sensory nucleus of trigeminal nerve development | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0008609 | alkylglycerone-phosphate synthase activity | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0008079 | translation termination factor activity | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0061141 | lung ciliated cell differentiation | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0042583 | chromaffin granule | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0030981 | cortical microtubule cytoskeleton | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0003747 | translation release factor activity | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 2 | 2 |
| GO:2000791 | negative regulation of mesenchymal cell proliferation involved in lung development | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0032286 | central nervous system myelin maintenance | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0051294 | establishment of spindle orientation | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:1900221 | regulation of beta-amyloid clearance | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:1902998 | positive regulation of neurofibrillary tangle assembly | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0008611 | ether lipid biosynthetic process | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0030529 | ribonucleoprotein complex | 1.89e-03 | 1.00e+00 | 4.896 | 2 | 5 | 82 |
| GO:0006366 | transcription from RNA polymerase II promoter | 2.07e-03 | 1.00e+00 | 3.425 | 3 | 23 | 341 |
| GO:0008762 | UDP-N-acetylmuramate dehydrogenase activity | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:1902004 | positive regulation of beta-amyloid formation | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:0017038 | protein import | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:0051788 | response to misfolded protein | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:0032463 | negative regulation of protein homooligomerization | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:0006260 | DNA replication | 2.47e-03 | 1.00e+00 | 4.699 | 2 | 9 | 94 |
| GO:0097452 | GAIT complex | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 2 | 4 |
| GO:0021960 | anterior commissure morphogenesis | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0006449 | regulation of translational termination | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0097418 | neurofibrillary tangle | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0044300 | cerebellar mossy fiber | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0060486 | Clara cell differentiation | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0043515 | kinetochore binding | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 2 | 4 |
| GO:0004521 | endoribonuclease activity | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0008610 | lipid biosynthetic process | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0010458 | exit from mitosis | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 2 | 5 |
| GO:0006398 | histone mRNA 3'-end processing | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0005828 | kinetochore microtubule | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 2 | 5 |
| GO:0060510 | Type II pneumocyte differentiation | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0021680 | cerebellar Purkinje cell layer development | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0031023 | microtubule organizing center organization | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0000398 | mRNA splicing, via spliceosome | 4.53e-03 | 1.00e+00 | 4.253 | 2 | 15 | 128 |
| GO:1902430 | negative regulation of beta-amyloid formation | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0021707 | cerebellar granule cell differentiation | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0071679 | commissural neuron axon guidance | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0097440 | apical dendrite | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0060509 | Type I pneumocyte differentiation | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 2 | 6 |
| GO:0008409 | 5'-3' exonuclease activity | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 2 | 7 |
| GO:0048708 | astrocyte differentiation | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 7 |
| GO:1901216 | positive regulation of neuron death | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 7 |
| GO:1901214 | regulation of neuron death | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 7 |
| GO:0034366 | spherical high-density lipoprotein particle | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 7 |
| GO:0051787 | misfolded protein binding | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 2 | 8 |
| GO:0071949 | FAD binding | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 1 | 8 |
| GO:0034453 | microtubule anchoring | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 9 |
| GO:0006379 | mRNA cleavage | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 2 | 9 |
| GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 2 | 9 |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 3 | 9 |
| GO:0001774 | microglial cell activation | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 9 |
| GO:0008380 | RNA splicing | 7.43e-03 | 1.00e+00 | 3.887 | 2 | 21 | 165 |
| GO:0008143 | poly(A) binding | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 2 | 10 |
| GO:0001578 | microtubule bundle formation | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 2 | 10 |
| GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 10 |
| GO:0051131 | chaperone-mediated protein complex assembly | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 3 | 10 |
| GO:0051010 | microtubule plus-end binding | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 11 |
| GO:0007026 | negative regulation of microtubule depolymerization | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 11 |
| GO:0007020 | microtubule nucleation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 11 |
| GO:0006479 | protein methylation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 11 |
| GO:0010001 | glial cell differentiation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 11 |
| GO:0030902 | hindbrain development | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 2 | 14 |
| GO:0043691 | reverse cholesterol transport | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 14 |
| GO:0005782 | peroxisomal matrix | 1.30e-02 | 1.00e+00 | 6.253 | 1 | 1 | 16 |
| GO:0043022 | ribosome binding | 1.38e-02 | 1.00e+00 | 6.166 | 1 | 2 | 17 |
| GO:0006378 | mRNA polyadenylation | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 3 | 18 |
| GO:0001106 | RNA polymerase II transcription corepressor activity | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 2 | 18 |
| GO:0006956 | complement activation | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 2 | 20 |
| GO:0007163 | establishment or maintenance of cell polarity | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 1 | 20 |
| GO:0071346 | cellular response to interferon-gamma | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 2 | 20 |
| GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1.71e-02 | 1.00e+00 | 5.861 | 1 | 2 | 21 |
| GO:0061077 | chaperone-mediated protein folding | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 22 |
| GO:0002062 | chondrocyte differentiation | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 2 | 22 |
| GO:0001836 | release of cytochrome c from mitochondria | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 22 |
| GO:0043392 | negative regulation of DNA binding | 1.87e-02 | 1.00e+00 | 5.730 | 1 | 1 | 23 |
| GO:0005881 | cytoplasmic microtubule | 1.95e-02 | 1.00e+00 | 5.668 | 1 | 3 | 24 |
| GO:0005876 | spindle microtubule | 1.95e-02 | 1.00e+00 | 5.668 | 1 | 4 | 24 |
| GO:0006958 | complement activation, classical pathway | 2.19e-02 | 1.00e+00 | 5.499 | 1 | 3 | 27 |
| GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 1 | 28 |
| GO:0005778 | peroxisomal membrane | 2.35e-02 | 1.00e+00 | 5.395 | 1 | 3 | 29 |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2.35e-02 | 1.00e+00 | 5.395 | 1 | 1 | 29 |
| GO:0032760 | positive regulation of tumor necrosis factor production | 2.59e-02 | 1.00e+00 | 5.253 | 1 | 1 | 32 |
| GO:0017148 | negative regulation of translation | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 5 | 33 |
| GO:0000902 | cell morphogenesis | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 5 | 35 |
| GO:0031093 | platelet alpha granule lumen | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 3 | 35 |
| GO:0031966 | mitochondrial membrane | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 1 | 35 |
| GO:0031124 | mRNA 3'-end processing | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 4 | 35 |
| GO:0000226 | microtubule cytoskeleton organization | 2.91e-02 | 1.00e+00 | 5.083 | 1 | 1 | 36 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 3.07e-02 | 1.00e+00 | 5.005 | 1 | 4 | 38 |
| GO:0006369 | termination of RNA polymerase II transcription | 3.07e-02 | 1.00e+00 | 5.005 | 1 | 5 | 38 |
| GO:0006396 | RNA processing | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 4 | 40 |
| GO:0006406 | mRNA export from nucleus | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 7 | 44 |
| GO:0005783 | endoplasmic reticulum | 3.64e-02 | 1.00e+00 | 2.680 | 2 | 12 | 381 |
| GO:0051301 | cell division | 3.95e-02 | 1.00e+00 | 4.639 | 1 | 2 | 49 |
| GO:0097193 | intrinsic apoptotic signaling pathway | 3.95e-02 | 1.00e+00 | 4.639 | 1 | 1 | 49 |
| GO:0005777 | peroxisome | 4.03e-02 | 1.00e+00 | 4.610 | 1 | 2 | 50 |
| GO:0000776 | kinetochore | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 8 | 55 |
| GO:0000777 | condensed chromosome kinetochore | 4.58e-02 | 1.00e+00 | 4.421 | 1 | 6 | 57 |
| GO:0003705 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity | 4.66e-02 | 1.00e+00 | 4.395 | 1 | 3 | 58 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 4.73e-02 | 1.00e+00 | 4.371 | 1 | 1 | 59 |
| GO:0071013 | catalytic step 2 spliceosome | 4.89e-02 | 1.00e+00 | 4.323 | 1 | 5 | 61 |
| GO:0002576 | platelet degranulation | 4.97e-02 | 1.00e+00 | 4.299 | 1 | 3 | 62 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 5.05e-02 | 1.00e+00 | 2.421 | 2 | 29 | 456 |
| GO:0006415 | translational termination | 5.05e-02 | 1.00e+00 | 4.276 | 1 | 2 | 63 |
| GO:0050821 | protein stabilization | 5.13e-02 | 1.00e+00 | 4.253 | 1 | 3 | 64 |
| GO:0006629 | lipid metabolic process | 5.20e-02 | 1.00e+00 | 4.231 | 1 | 2 | 65 |
| GO:0001649 | osteoblast differentiation | 5.91e-02 | 1.00e+00 | 4.044 | 1 | 5 | 74 |
| GO:0005938 | cell cortex | 6.14e-02 | 1.00e+00 | 3.987 | 1 | 6 | 77 |
| GO:0072562 | blood microparticle | 6.45e-02 | 1.00e+00 | 3.914 | 1 | 4 | 81 |
| GO:0016020 | membrane | 6.54e-02 | 1.00e+00 | 1.601 | 3 | 56 | 1207 |
| GO:0003690 | double-stranded DNA binding | 6.60e-02 | 1.00e+00 | 3.878 | 1 | 6 | 83 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6.68e-02 | 1.00e+00 | 3.861 | 1 | 5 | 84 |
| GO:0010467 | gene expression | 6.73e-02 | 1.00e+00 | 2.190 | 2 | 49 | 535 |
| GO:0044255 | cellular lipid metabolic process | 7.07e-02 | 1.00e+00 | 3.778 | 1 | 2 | 89 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 7.07e-02 | 1.00e+00 | 3.778 | 1 | 2 | 89 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 7.68e-02 | 1.00e+00 | 3.653 | 1 | 7 | 97 |
| GO:0008017 | microtubule binding | 7.68e-02 | 1.00e+00 | 3.653 | 1 | 9 | 97 |
| GO:0009615 | response to virus | 7.68e-02 | 1.00e+00 | 3.653 | 1 | 3 | 97 |
| GO:0016887 | ATPase activity | 7.91e-02 | 1.00e+00 | 3.610 | 1 | 5 | 100 |
| GO:0031012 | extracellular matrix | 8.44e-02 | 1.00e+00 | 3.512 | 1 | 4 | 107 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 9.33e-02 | 1.00e+00 | 1.922 | 2 | 29 | 644 |
| GO:0005739 | mitochondrion | 9.71e-02 | 1.00e+00 | 1.889 | 2 | 32 | 659 |
| GO:0001077 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 1.03e-01 | 1.00e+00 | 3.209 | 1 | 4 | 132 |
| GO:0031625 | ubiquitin protein ligase binding | 1.08e-01 | 1.00e+00 | 3.145 | 1 | 7 | 138 |
| GO:0030168 | platelet activation | 1.10e-01 | 1.00e+00 | 3.114 | 1 | 5 | 141 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.11e-01 | 1.00e+00 | 3.104 | 1 | 7 | 142 |
| GO:0016071 | mRNA metabolic process | 1.31e-01 | 1.00e+00 | 2.844 | 1 | 10 | 170 |
| GO:0044822 | poly(A) RNA binding | 1.35e-01 | 1.00e+00 | 1.611 | 2 | 45 | 799 |
| GO:0000166 | nucleotide binding | 1.35e-01 | 1.00e+00 | 2.794 | 1 | 13 | 176 |
| GO:0006412 | translation | 1.37e-01 | 1.00e+00 | 2.778 | 1 | 12 | 178 |
| GO:0016070 | RNA metabolic process | 1.44e-01 | 1.00e+00 | 2.699 | 1 | 10 | 188 |
| GO:0043065 | positive regulation of apoptotic process | 1.54e-01 | 1.00e+00 | 2.595 | 1 | 7 | 202 |
| GO:0005654 | nucleoplasm | 1.57e-01 | 1.00e+00 | 1.479 | 2 | 68 | 876 |
| GO:0043231 | intracellular membrane-bounded organelle | 1.65e-01 | 1.00e+00 | 2.485 | 1 | 8 | 218 |
| GO:0005813 | centrosome | 1.65e-01 | 1.00e+00 | 2.492 | 1 | 10 | 217 |
| GO:0005829 | cytosol | 1.67e-01 | 1.00e+00 | 1.035 | 3 | 88 | 1787 |
| GO:0003677 | DNA binding | 1.78e-01 | 1.00e+00 | 1.366 | 2 | 52 | 947 |
| GO:0007411 | axon guidance | 1.79e-01 | 1.00e+00 | 2.365 | 1 | 8 | 237 |
| GO:0005515 | protein binding | 2.10e-01 | 1.00e+00 | 0.566 | 5 | 198 | 4124 |
| GO:0055114 | oxidation-reduction process | 2.26e-01 | 1.00e+00 | 1.987 | 1 | 9 | 308 |
| GO:0000278 | mitotic cell cycle | 2.30e-01 | 1.00e+00 | 1.959 | 1 | 28 | 314 |
| GO:0007596 | blood coagulation | 2.46e-01 | 1.00e+00 | 1.848 | 1 | 13 | 339 |
| GO:0048471 | perinuclear region of cytoplasm | 2.49e-01 | 1.00e+00 | 1.827 | 1 | 11 | 344 |
| GO:0044267 | cellular protein metabolic process | 2.55e-01 | 1.00e+00 | 1.794 | 1 | 14 | 352 |
| GO:0005634 | nucleus | 2.57e-01 | 1.00e+00 | 0.589 | 4 | 158 | 3246 |
| GO:0005730 | nucleolus | 2.63e-01 | 1.00e+00 | 1.004 | 2 | 74 | 1217 |
| GO:0005794 | Golgi apparatus | 2.94e-01 | 1.00e+00 | 1.556 | 1 | 6 | 415 |
| GO:0016032 | viral process | 3.01e-01 | 1.00e+00 | 1.512 | 1 | 30 | 428 |
| GO:0045087 | innate immune response | 3.16e-01 | 1.00e+00 | 1.433 | 1 | 12 | 452 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 3.88e-01 | 1.00e+00 | 1.069 | 1 | 22 | 582 |
| GO:0070062 | extracellular vesicular exosome | 4.00e-01 | 1.00e+00 | 0.573 | 2 | 51 | 1641 |
| GO:0005615 | extracellular space | 4.24e-01 | 1.00e+00 | 0.907 | 1 | 15 | 651 |
| GO:0005576 | extracellular region | 4.30e-01 | 1.00e+00 | 0.881 | 1 | 15 | 663 |
| GO:0044281 | small molecule metabolic process | 5.15e-01 | 1.00e+00 | 0.532 | 1 | 35 | 844 |
| GO:0046872 | metal ion binding | 5.53e-01 | 1.00e+00 | 0.386 | 1 | 24 | 934 |
| GO:0005737 | cytoplasm | 6.80e-01 | 1.00e+00 | -0.109 | 2 | 127 | 2633 |