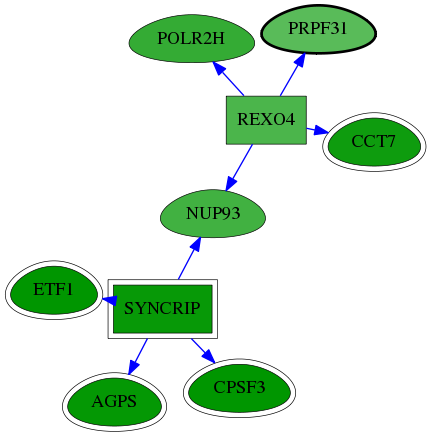
| GO:0000398 | mRNA splicing, via spliceosome | 3.37e-06 | 3.29e-02 | 5.083 | 4 | 15 | 128 |
| GO:0008380 | RNA splicing | 3.69e-04 | 1.00e+00 | 4.302 | 3 | 21 | 165 |
| GO:0008609 | alkylglycerone-phosphate synthase activity | 9.22e-04 | 1.00e+00 | 10.083 | 1 | 1 | 1 |
| GO:0016149 | translation release factor activity, codon specific | 9.22e-04 | 1.00e+00 | 10.083 | 1 | 1 | 1 |
| GO:0008079 | translation termination factor activity | 9.22e-04 | 1.00e+00 | 10.083 | 1 | 1 | 1 |
| GO:0070990 | snRNP binding | 9.22e-04 | 1.00e+00 | 10.083 | 1 | 1 | 1 |
| GO:0005690 | U4atac snRNP | 9.22e-04 | 1.00e+00 | 10.083 | 1 | 1 | 1 |
| GO:0003723 | RNA binding | 1.20e-03 | 1.00e+00 | 3.720 | 3 | 22 | 247 |
| GO:0005684 | U2-type spliceosomal complex | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 2 | 2 |
| GO:0003747 | translation release factor activity | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 2 | 2 |
| GO:0008611 | ether lipid biosynthetic process | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 2 |
| GO:0030529 | ribonucleoprotein complex | 2.41e-03 | 1.00e+00 | 4.726 | 2 | 5 | 82 |
| GO:0008762 | UDP-N-acetylmuramate dehydrogenase activity | 2.76e-03 | 1.00e+00 | 8.499 | 1 | 1 | 3 |
| GO:0006449 | regulation of translational termination | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 4 |
| GO:0097452 | GAIT complex | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 2 | 4 |
| GO:0046540 | U4/U6 x U5 tri-snRNP complex | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 4 |
| GO:0044822 | poly(A) RNA binding | 4.01e-03 | 1.00e+00 | 2.441 | 4 | 45 | 799 |
| GO:0004521 | endoribonuclease activity | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 5 |
| GO:0008610 | lipid biosynthetic process | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 5 |
| GO:0006398 | histone mRNA 3'-end processing | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 5 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 2 | 5 |
| GO:0070937 | CRD-mediated mRNA stability complex | 5.52e-03 | 1.00e+00 | 7.499 | 1 | 2 | 6 |
| GO:0001055 | RNA polymerase II activity | 5.52e-03 | 1.00e+00 | 7.499 | 1 | 3 | 6 |
| GO:0016032 | viral process | 5.76e-03 | 1.00e+00 | 2.927 | 3 | 30 | 428 |
| GO:0002199 | zona pellucida receptor complex | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 4 | 7 |
| GO:0005832 | chaperonin-containing T-complex | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 4 | 7 |
| GO:0008409 | 5'-3' exonuclease activity | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 2 | 7 |
| GO:0071949 | FAD binding | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 8 |
| GO:0051292 | nuclear pore complex assembly | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 4 | 8 |
| GO:0017056 | structural constituent of nuclear pore | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 3 | 8 |
| GO:0006379 | mRNA cleavage | 8.27e-03 | 1.00e+00 | 6.914 | 1 | 2 | 9 |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 8.27e-03 | 1.00e+00 | 6.914 | 1 | 3 | 9 |
| GO:0000244 | spliceosomal tri-snRNP complex assembly | 8.27e-03 | 1.00e+00 | 6.914 | 1 | 1 | 9 |
| GO:0005687 | U4 snRNP | 8.27e-03 | 1.00e+00 | 6.914 | 1 | 1 | 9 |
| GO:0005736 | DNA-directed RNA polymerase I complex | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 4 | 10 |
| GO:0001054 | RNA polymerase I activity | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 4 | 10 |
| GO:0008143 | poly(A) binding | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 2 | 10 |
| GO:0006479 | protein methylation | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 2 | 11 |
| GO:0010467 | gene expression | 1.07e-02 | 1.00e+00 | 2.605 | 3 | 49 | 535 |
| GO:0004527 | exonuclease activity | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 4 | 12 |
| GO:0034399 | nuclear periphery | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 3 | 12 |
| GO:0005665 | DNA-directed RNA polymerase II, core complex | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 3 | 13 |
| GO:0001056 | RNA polymerase III activity | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 3 | 13 |
| GO:0005666 | DNA-directed RNA polymerase III complex | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 3 | 13 |
| GO:0006385 | transcription elongation from RNA polymerase III promoter | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 3 | 14 |
| GO:0006386 | termination of RNA polymerase III transcription | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 3 | 14 |
| GO:0007339 | binding of sperm to zona pellucida | 1.37e-02 | 1.00e+00 | 6.177 | 1 | 4 | 15 |
| GO:0005782 | peroxisomal matrix | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 1 | 16 |
| GO:0043021 | ribonucleoprotein complex binding | 1.56e-02 | 1.00e+00 | 5.996 | 1 | 1 | 17 |
| GO:0043022 | ribosome binding | 1.56e-02 | 1.00e+00 | 5.996 | 1 | 2 | 17 |
| GO:0071339 | MLL1 complex | 1.65e-02 | 1.00e+00 | 5.914 | 1 | 2 | 18 |
| GO:0006378 | mRNA polyadenylation | 1.65e-02 | 1.00e+00 | 5.914 | 1 | 3 | 18 |
| GO:0071346 | cellular response to interferon-gamma | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 2 | 20 |
| GO:0006362 | transcription elongation from RNA polymerase I promoter | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 3 | 20 |
| GO:0006363 | termination of RNA polymerase I transcription | 1.92e-02 | 1.00e+00 | 5.691 | 1 | 3 | 21 |
| GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1.92e-02 | 1.00e+00 | 5.691 | 1 | 2 | 21 |
| GO:0051084 | 'de novo' posttranslational protein folding | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 4 | 22 |
| GO:0006361 | transcription initiation from RNA polymerase I promoter | 2.10e-02 | 1.00e+00 | 5.560 | 1 | 2 | 23 |
| GO:0006370 | 7-methylguanosine mRNA capping | 2.19e-02 | 1.00e+00 | 5.499 | 1 | 5 | 24 |
| GO:0010827 | regulation of glucose transport | 2.28e-02 | 1.00e+00 | 5.440 | 1 | 7 | 25 |
| GO:0006360 | transcription from RNA polymerase I promoter | 2.46e-02 | 1.00e+00 | 5.329 | 1 | 4 | 27 |
| GO:0005778 | peroxisomal membrane | 2.64e-02 | 1.00e+00 | 5.225 | 1 | 3 | 29 |
| GO:0006383 | transcription from RNA polymerase III promoter | 2.64e-02 | 1.00e+00 | 5.225 | 1 | 6 | 29 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 2.82e-02 | 1.00e+00 | 5.129 | 1 | 6 | 31 |
| GO:0007077 | mitotic nuclear envelope disassembly | 2.91e-02 | 1.00e+00 | 5.083 | 1 | 9 | 32 |
| GO:0015030 | Cajal body | 2.91e-02 | 1.00e+00 | 5.083 | 1 | 3 | 32 |
| GO:0017148 | negative regulation of translation | 3.00e-02 | 1.00e+00 | 5.039 | 1 | 5 | 33 |
| GO:0050434 | positive regulation of viral transcription | 3.18e-02 | 1.00e+00 | 4.954 | 1 | 5 | 35 |
| GO:0031124 | mRNA 3'-end processing | 3.18e-02 | 1.00e+00 | 4.954 | 1 | 4 | 35 |
| GO:0044297 | cell body | 3.18e-02 | 1.00e+00 | 4.954 | 1 | 5 | 35 |
| GO:0008645 | hexose transport | 3.27e-02 | 1.00e+00 | 4.914 | 1 | 8 | 36 |
| GO:0006369 | termination of RNA polymerase II transcription | 3.45e-02 | 1.00e+00 | 4.836 | 1 | 5 | 38 |
| GO:0006396 | RNA processing | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 4 | 40 |
| GO:0006283 | transcription-coupled nucleotide-excision repair | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 6 | 40 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.72e-02 | 1.00e+00 | 2.670 | 2 | 23 | 341 |
| GO:0044267 | cellular protein metabolic process | 3.94e-02 | 1.00e+00 | 2.624 | 2 | 14 | 352 |
| GO:0006406 | mRNA export from nucleus | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 7 | 44 |
| GO:0005654 | nucleoplasm | 4.01e-02 | 1.00e+00 | 1.894 | 3 | 68 | 876 |
| GO:0051028 | mRNA transport | 4.25e-02 | 1.00e+00 | 4.529 | 1 | 5 | 47 |
| GO:0032481 | positive regulation of type I interferon production | 4.34e-02 | 1.00e+00 | 4.499 | 1 | 3 | 48 |
| GO:0015758 | glucose transport | 4.34e-02 | 1.00e+00 | 4.499 | 1 | 8 | 48 |
| GO:0005777 | peroxisome | 4.52e-02 | 1.00e+00 | 4.440 | 1 | 2 | 50 |
| GO:0005643 | nuclear pore | 4.61e-02 | 1.00e+00 | 4.411 | 1 | 12 | 51 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 4.69e-02 | 1.00e+00 | 4.383 | 1 | 8 | 52 |
| GO:0006289 | nucleotide-excision repair | 4.78e-02 | 1.00e+00 | 4.356 | 1 | 6 | 53 |
| GO:0071013 | catalytic step 2 spliceosome | 5.49e-02 | 1.00e+00 | 4.153 | 1 | 5 | 61 |
| GO:0006415 | translational termination | 5.66e-02 | 1.00e+00 | 4.106 | 1 | 2 | 63 |
| GO:0051082 | unfolded protein binding | 6.27e-02 | 1.00e+00 | 3.954 | 1 | 5 | 70 |
| GO:0001649 | osteoblast differentiation | 6.62e-02 | 1.00e+00 | 3.874 | 1 | 5 | 74 |
| GO:0005635 | nuclear envelope | 7.40e-02 | 1.00e+00 | 3.708 | 1 | 6 | 83 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 7.48e-02 | 1.00e+00 | 3.691 | 1 | 5 | 84 |
| GO:0044255 | cellular lipid metabolic process | 7.91e-02 | 1.00e+00 | 3.608 | 1 | 2 | 89 |
| GO:0006457 | protein folding | 8.60e-02 | 1.00e+00 | 3.484 | 1 | 5 | 97 |
| GO:0016020 | membrane | 8.94e-02 | 1.00e+00 | 1.431 | 3 | 56 | 1207 |
| GO:0016607 | nuclear speck | 1.07e-01 | 1.00e+00 | 3.153 | 1 | 8 | 122 |
| GO:0031965 | nuclear membrane | 1.14e-01 | 1.00e+00 | 3.061 | 1 | 8 | 130 |
| GO:0005739 | mitochondrion | 1.19e-01 | 1.00e+00 | 1.719 | 2 | 32 | 659 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.39e-01 | 1.00e+00 | 2.753 | 1 | 18 | 161 |
| GO:0005874 | microtubule | 1.41e-01 | 1.00e+00 | 2.735 | 1 | 10 | 163 |
| GO:0016071 | mRNA metabolic process | 1.46e-01 | 1.00e+00 | 2.674 | 1 | 10 | 170 |
| GO:0019221 | cytokine-mediated signaling pathway | 1.48e-01 | 1.00e+00 | 2.657 | 1 | 10 | 172 |
| GO:0000166 | nucleotide binding | 1.51e-01 | 1.00e+00 | 2.624 | 1 | 13 | 176 |
| GO:0006412 | translation | 1.53e-01 | 1.00e+00 | 2.608 | 1 | 12 | 178 |
| GO:0005975 | carbohydrate metabolic process | 1.59e-01 | 1.00e+00 | 2.544 | 1 | 10 | 186 |
| GO:0016070 | RNA metabolic process | 1.61e-01 | 1.00e+00 | 2.529 | 1 | 10 | 188 |
| GO:0015031 | protein transport | 1.72e-01 | 1.00e+00 | 2.418 | 1 | 10 | 203 |
| GO:0006281 | DNA repair | 1.72e-01 | 1.00e+00 | 2.418 | 1 | 24 | 203 |
| GO:0044281 | small molecule metabolic process | 1.79e-01 | 1.00e+00 | 1.362 | 2 | 35 | 844 |
| GO:0043231 | intracellular membrane-bounded organelle | 1.84e-01 | 1.00e+00 | 2.315 | 1 | 8 | 218 |
| GO:0005829 | cytosol | 2.18e-01 | 1.00e+00 | 0.865 | 3 | 88 | 1787 |
| GO:0055114 | oxidation-reduction process | 2.51e-01 | 1.00e+00 | 1.817 | 1 | 9 | 308 |
| GO:0000278 | mitotic cell cycle | 2.55e-01 | 1.00e+00 | 1.789 | 1 | 28 | 314 |
| GO:0055085 | transmembrane transport | 2.66e-01 | 1.00e+00 | 1.722 | 1 | 13 | 329 |
| GO:0042802 | identical protein binding | 2.83e-01 | 1.00e+00 | 1.616 | 1 | 15 | 354 |
| GO:0005783 | endoplasmic reticulum | 3.01e-01 | 1.00e+00 | 1.510 | 1 | 12 | 381 |
| GO:0005730 | nucleolus | 3.11e-01 | 1.00e+00 | 0.834 | 2 | 74 | 1217 |
| GO:0005515 | protein binding | 3.15e-01 | 1.00e+00 | 0.396 | 5 | 198 | 4124 |
| GO:0045087 | innate immune response | 3.47e-01 | 1.00e+00 | 1.263 | 1 | 12 | 452 |
| GO:0005634 | nucleus | 3.47e-01 | 1.00e+00 | 0.419 | 4 | 158 | 3246 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 4.25e-01 | 1.00e+00 | 0.899 | 1 | 22 | 582 |
| GO:0006355 | regulation of transcription, DNA-templated | 4.96e-01 | 1.00e+00 | 0.602 | 1 | 31 | 715 |
| GO:0005524 | ATP binding | 5.78e-01 | 1.00e+00 | 0.283 | 1 | 37 | 892 |
| GO:0046872 | metal ion binding | 5.96e-01 | 1.00e+00 | 0.216 | 1 | 24 | 934 |
| GO:0003677 | DNA binding | 6.01e-01 | 1.00e+00 | 0.196 | 1 | 52 | 947 |
| GO:0005737 | cytoplasm | 7.45e-01 | 1.00e+00 | -0.279 | 2 | 127 | 2633 |
| GO:0070062 | extracellular vesicular exosome | 8.09e-01 | 1.00e+00 | -0.597 | 1 | 51 | 1641 |