int-snw-79711
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.842 | 1.39e-15 | 4.44e-04 | 3.19e-03 |
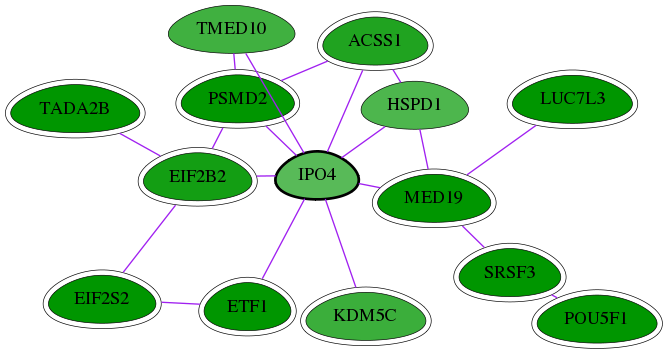
Genes (14)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| PSMD2 | 5708 | 47 | -4.172 | 3.157 | 386 | Yes | Yes |
| MED19 | 219541 | 30 | -2.657 | 3.449 | 115 | Yes | - |
| EIF2S2 | 8894 | 9 | -2.999 | 2.968 | 81 | Yes | Yes |
| TMED10 | 10972 | 3 | -1.992 | 3.104 | 106 | - | - |
| LUC7L3 | 51747 | 33 | -3.523 | 3.449 | 18 | Yes | - |
| TADA2B | 93624 | 11 | -3.061 | 3.082 | 19 | Yes | - |
| EIF2B2 | 8892 | 8 | -2.462 | 2.968 | 95 | Yes | Yes |
| ETF1 | 2107 | 3 | -2.829 | 2.959 | 71 | Yes | Yes |
| KDM5C | 8242 | 2 | -2.040 | 2.842 | 45 | Yes | - |
| SRSF3 | 6428 | 73 | -2.992 | 3.538 | 54 | Yes | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| HSPD1 | 3329 | 3 | -1.852 | 2.870 | 286 | - | - |
| ACSS1 | 84532 | 2 | -2.326 | 2.842 | 148 | Yes | - |
| [ IPO4 ] | 79711 | 2 | -1.739 | 2.842 | 177 | - | - |
Interactions (19)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| EIF2B2 | 8892 | TADA2B | 93624 | pp | -- | int.I2D: BioGrid_Fly, BIND_Fly, FlyLow, IntAct_Fly, MINT_Fly |
| TMED10 | 10972 | IPO4 | 79711 | pp | -- | int.I2D: IntAct_Yeast, YeastLow |
| SRSF3 | 6428 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| EIF2B2 | 8892 | IPO4 | 79711 | pp | -- | int.I2D: IntAct_Yeast |
| HSPD1 | 3329 | ACSS1 | 84532 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| ETF1 | 2107 | IPO4 | 79711 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastLow |
| IPO4 | 79711 | ACSS1 | 84532 | pp | -- | int.I2D: IntAct_Yeast |
| PSMD2 | 5708 | IPO4 | 79711 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMD2 | 5708 | TMED10 | 10972 | pp | -- | int.I2D: IntAct_Yeast |
| IPO4 | 79711 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| PSMD2 | 5708 | EIF2B2 | 8892 | pp | -- | int.I2D: IntAct_Yeast |
| POU5F1 | 5460 | SRSF3 | 6428 | pp | -- | int.I2D: BioGrid |
| LUC7L3 | 51747 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| PSMD2 | 5708 | ACSS1 | 84532 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| KDM5C | 8242 | IPO4 | 79711 | pp | -- | int.I2D: YeastLow |
| HSPD1 | 3329 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| EIF2B2 | 8892 | EIF2S2 | 8894 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh |
| HSPD1 | 3329 | IPO4 | 79711 | pp | -- | int.I2D: YeastLow |
| ETF1 | 2107 | EIF2S2 | 8894 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
Related GO terms (214)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0044822 | poly(A) RNA binding | 2.73e-04 | 1.00e+00 | 2.549 | 6 | 50 | 1056 |
| GO:0010467 | gene expression | 2.98e-04 | 1.00e+00 | 2.945 | 5 | 45 | 669 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0048199 | vesicle targeting, to, from or within Golgi | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0035964 | COPI-coated vesicle budding | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0006085 | acetyl-CoA biosynthetic process | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0008079 | translation termination factor activity | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0048291 | isotype switching to IgG isotypes | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:1902003 | regulation of beta-amyloid formation | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0002368 | B cell cytokine production | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0003743 | translation initiation factor activity | 1.00e-03 | 1.00e+00 | 5.394 | 2 | 7 | 49 |
| GO:0006412 | translation | 1.28e-03 | 1.00e+00 | 3.748 | 3 | 8 | 230 |
| GO:0070765 | gamma-secretase complex | 1.94e-03 | 1.00e+00 | 9.009 | 1 | 1 | 2 |
| GO:0019542 | propionate biosynthetic process | 1.94e-03 | 1.00e+00 | 9.009 | 1 | 1 | 2 |
| GO:0002176 | male germ cell proliferation | 1.94e-03 | 1.00e+00 | 9.009 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.94e-03 | 1.00e+00 | 9.009 | 1 | 1 | 2 |
| GO:0019427 | acetyl-CoA biosynthetic process from acetate | 1.94e-03 | 1.00e+00 | 9.009 | 1 | 1 | 2 |
| GO:0019413 | acetate biosynthetic process | 1.94e-03 | 1.00e+00 | 9.009 | 1 | 1 | 2 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0006458 | 'de novo' protein folding | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0016149 | translation release factor activity, codon specific | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0003987 | acetate-CoA ligase activity | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0030135 | coated vesicle | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0035459 | cargo loading into vesicle | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 1 | 4 |
| GO:0051716 | cellular response to stimulus | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 4 | 4 |
| GO:0046523 | S-methyl-5-thioribose-1-phosphate isomerase activity | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 3 | 4 |
| GO:0030137 | COPI-coated vesicle | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 1 | 4 |
| GO:0003747 | translation release factor activity | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 2 | 4 |
| GO:0032453 | histone demethylase activity (H3-K4 specific) | 3.88e-03 | 1.00e+00 | 8.009 | 1 | 1 | 4 |
| GO:0003723 | RNA binding | 3.96e-03 | 1.00e+00 | 3.176 | 3 | 18 | 342 |
| GO:0070461 | SAGA-type complex | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 5 |
| GO:0001714 | endodermal cell fate specification | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 5 |
| GO:0046696 | lipopolysaccharide receptor complex | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 5 |
| GO:0043279 | response to alkaloid | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 5 |
| GO:0006449 | regulation of translational termination | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 5 |
| GO:0003688 | DNA replication origin binding | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 6 |
| GO:0034720 | histone H3-K4 demethylation | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 6 |
| GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 6 |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 4 | 6 |
| GO:0006413 | translational initiation | 6.94e-03 | 1.00e+00 | 3.975 | 2 | 7 | 131 |
| GO:0006366 | transcription from RNA polymerase II promoter | 6.98e-03 | 1.00e+00 | 2.883 | 3 | 30 | 419 |
| GO:0051604 | protein maturation | 7.74e-03 | 1.00e+00 | 7.009 | 1 | 2 | 8 |
| GO:0016208 | AMP binding | 8.70e-03 | 1.00e+00 | 6.839 | 1 | 1 | 9 |
| GO:0019509 | L-methionine biosynthetic process from methylthioadenosine | 8.70e-03 | 1.00e+00 | 6.839 | 1 | 3 | 9 |
| GO:0035413 | positive regulation of catenin import into nucleus | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 2 | 10 |
| GO:0045055 | regulated secretory pathway | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 10 |
| GO:0042589 | zymogen granule membrane | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 10 |
| GO:0006069 | ethanol oxidation | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 10 |
| GO:0043032 | positive regulation of macrophage activation | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 10 |
| GO:0044267 | cellular protein metabolic process | 9.80e-03 | 1.00e+00 | 2.705 | 3 | 20 | 474 |
| GO:0032727 | positive regulation of interferon-alpha production | 1.06e-02 | 1.00e+00 | 6.549 | 1 | 1 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 1.06e-02 | 1.00e+00 | 6.549 | 1 | 4 | 11 |
| GO:0030667 | secretory granule membrane | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 2 | 12 |
| GO:0030140 | trans-Golgi network transport vesicle | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 1 | 12 |
| GO:0005838 | proteasome regulatory particle | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 1 | 12 |
| GO:0006886 | intracellular protein transport | 1.16e-02 | 1.00e+00 | 3.591 | 2 | 4 | 171 |
| GO:0035198 | miRNA binding | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 1 | 12 |
| GO:0016607 | nuclear speck | 1.17e-02 | 1.00e+00 | 3.582 | 2 | 12 | 172 |
| GO:0048205 | COPI coating of Golgi vesicle | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 2 | 13 |
| GO:0030914 | STAGA complex | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 3 | 13 |
| GO:0030234 | enzyme regulator activity | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 2 | 13 |
| GO:0001530 | lipopolysaccharide binding | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 13 |
| GO:0051131 | chaperone-mediated protein complex assembly | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 13 |
| GO:0006479 | protein methylation | 1.35e-02 | 1.00e+00 | 6.201 | 1 | 2 | 14 |
| GO:0005685 | U1 snRNP | 1.35e-02 | 1.00e+00 | 6.201 | 1 | 1 | 14 |
| GO:0048208 | COPII vesicle coating | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 15 |
| GO:0001824 | blastocyst development | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 2 | 15 |
| GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 15 |
| GO:0042026 | protein refolding | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 15 |
| GO:0006376 | mRNA splice site selection | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 15 |
| GO:0043274 | phospholipase binding | 1.54e-02 | 1.00e+00 | 6.009 | 1 | 1 | 16 |
| GO:0042176 | regulation of protein catabolic process | 1.54e-02 | 1.00e+00 | 6.009 | 1 | 2 | 16 |
| GO:0006336 | DNA replication-independent nucleosome assembly | 1.54e-02 | 1.00e+00 | 6.009 | 1 | 2 | 16 |
| GO:0050870 | positive regulation of T cell activation | 1.64e-02 | 1.00e+00 | 5.921 | 1 | 1 | 17 |
| GO:0022624 | proteasome accessory complex | 1.64e-02 | 1.00e+00 | 5.921 | 1 | 1 | 17 |
| GO:0006335 | DNA replication-dependent nucleosome assembly | 1.64e-02 | 1.00e+00 | 5.921 | 1 | 2 | 17 |
| GO:0032733 | positive regulation of interleukin-10 production | 1.83e-02 | 1.00e+00 | 5.761 | 1 | 2 | 19 |
| GO:0016071 | mRNA metabolic process | 1.92e-02 | 1.00e+00 | 3.208 | 2 | 8 | 223 |
| GO:0014003 | oligodendrocyte development | 1.92e-02 | 1.00e+00 | 5.687 | 1 | 5 | 20 |
| GO:0008380 | RNA splicing | 2.00e-02 | 1.00e+00 | 3.176 | 2 | 22 | 228 |
| GO:0005759 | mitochondrial matrix | 2.00e-02 | 1.00e+00 | 3.176 | 2 | 4 | 228 |
| GO:0042100 | B cell proliferation | 2.02e-02 | 1.00e+00 | 5.616 | 1 | 1 | 21 |
| GO:0005829 | cytosol | 2.27e-02 | 1.00e+00 | 1.308 | 6 | 86 | 2496 |
| GO:0008135 | translation factor activity, nucleic acid binding | 2.31e-02 | 1.00e+00 | 5.424 | 1 | 3 | 24 |
| GO:0016070 | RNA metabolic process | 2.32e-02 | 1.00e+00 | 3.060 | 2 | 8 | 247 |
| GO:0008536 | Ran GTPase binding | 2.40e-02 | 1.00e+00 | 5.365 | 1 | 2 | 25 |
| GO:0042113 | B cell activation | 2.40e-02 | 1.00e+00 | 5.365 | 1 | 2 | 25 |
| GO:0032735 | positive regulation of interleukin-12 production | 2.40e-02 | 1.00e+00 | 5.365 | 1 | 2 | 25 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 2.40e-02 | 1.00e+00 | 5.365 | 1 | 1 | 25 |
| GO:0005515 | protein binding | 2.44e-02 | 1.00e+00 | 0.774 | 10 | 198 | 6024 |
| GO:0003677 | DNA binding | 2.55e-02 | 1.00e+00 | 1.758 | 4 | 49 | 1218 |
| GO:0043022 | ribosome binding | 2.59e-02 | 1.00e+00 | 5.254 | 1 | 1 | 27 |
| GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane | 2.59e-02 | 1.00e+00 | 5.254 | 1 | 1 | 27 |
| GO:0030658 | transport vesicle membrane | 3.06e-02 | 1.00e+00 | 5.009 | 1 | 1 | 32 |
| GO:0019905 | syntaxin binding | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 2 | 33 |
| GO:0001104 | RNA polymerase II transcription cofactor activity | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 8 | 33 |
| GO:0005801 | cis-Golgi network | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 1 | 33 |
| GO:0043234 | protein complex | 3.23e-02 | 1.00e+00 | 2.804 | 2 | 11 | 295 |
| GO:0042552 | myelination | 3.35e-02 | 1.00e+00 | 4.879 | 1 | 4 | 35 |
| GO:0016592 | mediator complex | 3.35e-02 | 1.00e+00 | 4.879 | 1 | 10 | 35 |
| GO:0032755 | positive regulation of interleukin-6 production | 3.44e-02 | 1.00e+00 | 4.839 | 1 | 2 | 36 |
| GO:0006446 | regulation of translational initiation | 3.44e-02 | 1.00e+00 | 4.839 | 1 | 4 | 36 |
| GO:0001541 | ovarian follicle development | 3.53e-02 | 1.00e+00 | 4.799 | 1 | 2 | 37 |
| GO:0032729 | positive regulation of interferon-gamma production | 3.72e-02 | 1.00e+00 | 4.723 | 1 | 1 | 39 |
| GO:0051259 | protein oligomerization | 3.72e-02 | 1.00e+00 | 4.723 | 1 | 2 | 39 |
| GO:0009408 | response to heat | 3.82e-02 | 1.00e+00 | 4.687 | 1 | 6 | 40 |
| GO:0035019 | somatic stem cell maintenance | 3.82e-02 | 1.00e+00 | 4.687 | 1 | 6 | 40 |
| GO:0042110 | T cell activation | 4.00e-02 | 1.00e+00 | 4.616 | 1 | 1 | 42 |
| GO:0031124 | mRNA 3'-end processing | 4.00e-02 | 1.00e+00 | 4.616 | 1 | 2 | 42 |
| GO:0043434 | response to peptide hormone | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 5 | 45 |
| GO:0006369 | termination of RNA polymerase II transcription | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 2 | 45 |
| GO:0048511 | rhythmic process | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 4 | 45 |
| GO:0007030 | Golgi organization | 4.65e-02 | 1.00e+00 | 4.394 | 1 | 3 | 49 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 4.75e-02 | 1.00e+00 | 4.365 | 1 | 1 | 50 |
| GO:0006986 | response to unfolded protein | 4.75e-02 | 1.00e+00 | 4.365 | 1 | 4 | 50 |
| GO:0005905 | coated pit | 4.75e-02 | 1.00e+00 | 4.365 | 1 | 6 | 50 |
| GO:0003725 | double-stranded RNA binding | 4.93e-02 | 1.00e+00 | 4.308 | 1 | 1 | 52 |
| GO:0009611 | response to wounding | 4.93e-02 | 1.00e+00 | 4.308 | 1 | 5 | 52 |
| GO:0006888 | ER to Golgi vesicle-mediated transport | 5.03e-02 | 1.00e+00 | 4.281 | 1 | 1 | 53 |
| GO:0002039 | p53 binding | 5.21e-02 | 1.00e+00 | 4.227 | 1 | 4 | 55 |
| GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment | 5.30e-02 | 1.00e+00 | 4.201 | 1 | 2 | 56 |
| GO:0005643 | nuclear pore | 5.40e-02 | 1.00e+00 | 4.176 | 1 | 11 | 57 |
| GO:0000502 | proteasome complex | 5.49e-02 | 1.00e+00 | 4.151 | 1 | 3 | 58 |
| GO:0051087 | chaperone binding | 5.58e-02 | 1.00e+00 | 4.126 | 1 | 3 | 59 |
| GO:0006406 | mRNA export from nucleus | 5.76e-02 | 1.00e+00 | 4.078 | 1 | 4 | 61 |
| GO:0030141 | secretory granule | 5.86e-02 | 1.00e+00 | 4.055 | 1 | 4 | 62 |
| GO:0009749 | response to glucose | 6.04e-02 | 1.00e+00 | 4.009 | 1 | 7 | 64 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 6.04e-02 | 1.00e+00 | 4.009 | 1 | 1 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 6.13e-02 | 1.00e+00 | 3.986 | 1 | 2 | 65 |
| GO:0043066 | negative regulation of apoptotic process | 6.21e-02 | 1.00e+00 | 2.281 | 2 | 16 | 424 |
| GO:0010468 | regulation of gene expression | 6.40e-02 | 1.00e+00 | 3.921 | 1 | 2 | 68 |
| GO:0003697 | single-stranded DNA binding | 6.40e-02 | 1.00e+00 | 3.921 | 1 | 5 | 68 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 6.59e-02 | 1.00e+00 | 3.879 | 1 | 2 | 70 |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 6.68e-02 | 1.00e+00 | 3.859 | 1 | 5 | 71 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 6.77e-02 | 1.00e+00 | 3.839 | 1 | 1 | 72 |
| GO:0003729 | mRNA binding | 6.77e-02 | 1.00e+00 | 3.839 | 1 | 4 | 72 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 6.95e-02 | 1.00e+00 | 3.799 | 1 | 4 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 6.95e-02 | 1.00e+00 | 3.799 | 1 | 2 | 74 |
| GO:0008584 | male gonad development | 7.04e-02 | 1.00e+00 | 3.780 | 1 | 3 | 75 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 7.04e-02 | 1.00e+00 | 3.780 | 1 | 1 | 75 |
| GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 7.22e-02 | 1.00e+00 | 3.742 | 1 | 3 | 77 |
| GO:0001822 | kidney development | 7.31e-02 | 1.00e+00 | 3.723 | 1 | 3 | 78 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 7.40e-02 | 1.00e+00 | 3.705 | 1 | 2 | 79 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 7.49e-02 | 1.00e+00 | 3.687 | 1 | 5 | 80 |
| GO:0009653 | anatomical structure morphogenesis | 7.77e-02 | 1.00e+00 | 3.634 | 1 | 2 | 83 |
| GO:0006415 | translational termination | 8.13e-02 | 1.00e+00 | 3.566 | 1 | 2 | 87 |
| GO:0050821 | protein stabilization | 8.21e-02 | 1.00e+00 | 3.549 | 1 | 3 | 88 |
| GO:0005654 | nucleoplasm | 8.23e-02 | 1.00e+00 | 1.514 | 3 | 64 | 1082 |
| GO:0042470 | melanosome | 8.48e-02 | 1.00e+00 | 3.501 | 1 | 2 | 91 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 8.57e-02 | 1.00e+00 | 3.485 | 1 | 1 | 92 |
| GO:0051082 | unfolded protein binding | 8.66e-02 | 1.00e+00 | 3.470 | 1 | 7 | 93 |
| GO:0007417 | central nervous system development | 8.93e-02 | 1.00e+00 | 3.424 | 1 | 5 | 96 |
| GO:0016032 | viral process | 9.28e-02 | 1.00e+00 | 1.948 | 2 | 26 | 534 |
| GO:0006805 | xenobiotic metabolic process | 9.55e-02 | 1.00e+00 | 3.322 | 1 | 2 | 103 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1.05e-01 | 1.00e+00 | 3.176 | 1 | 4 | 114 |
| GO:0000209 | protein polyubiquitination | 1.07e-01 | 1.00e+00 | 3.151 | 1 | 3 | 116 |
| GO:0006325 | chromatin organization | 1.09e-01 | 1.00e+00 | 3.126 | 1 | 12 | 118 |
| GO:0000790 | nuclear chromatin | 1.18e-01 | 1.00e+00 | 2.998 | 1 | 6 | 129 |
| GO:0005524 | ATP binding | 1.25e-01 | 1.00e+00 | 1.252 | 3 | 31 | 1298 |
| GO:0016887 | ATPase activity | 1.28e-01 | 1.00e+00 | 2.879 | 1 | 1 | 140 |
| GO:0005737 | cytoplasm | 1.32e-01 | 1.00e+00 | 0.714 | 6 | 124 | 3767 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.35e-01 | 1.00e+00 | 2.790 | 1 | 3 | 149 |
| GO:0042981 | regulation of apoptotic process | 1.36e-01 | 1.00e+00 | 2.780 | 1 | 4 | 150 |
| GO:0005769 | early endosome | 1.38e-01 | 1.00e+00 | 2.761 | 1 | 1 | 152 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.48e-01 | 1.00e+00 | 2.651 | 1 | 15 | 164 |
| GO:0044212 | transcription regulatory region DNA binding | 1.51e-01 | 1.00e+00 | 2.616 | 1 | 17 | 168 |
| GO:0034641 | cellular nitrogen compound metabolic process | 1.54e-01 | 1.00e+00 | 2.591 | 1 | 2 | 171 |
| GO:0005667 | transcription factor complex | 1.57e-01 | 1.00e+00 | 2.558 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.60e-01 | 1.00e+00 | 2.533 | 1 | 5 | 178 |
| GO:0043547 | positive regulation of GTPase activity | 1.61e-01 | 1.00e+00 | 2.517 | 1 | 9 | 180 |
| GO:0032403 | protein complex binding | 1.64e-01 | 1.00e+00 | 2.493 | 1 | 6 | 183 |
| GO:0001701 | in utero embryonic development | 1.78e-01 | 1.00e+00 | 2.358 | 1 | 8 | 201 |
| GO:0008134 | transcription factor binding | 2.11e-01 | 1.00e+00 | 2.090 | 1 | 18 | 242 |
| GO:0016020 | membrane | 2.18e-01 | 1.00e+00 | 0.879 | 3 | 46 | 1681 |
| GO:0000166 | nucleotide binding | 2.24e-01 | 1.00e+00 | 1.992 | 1 | 13 | 259 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 2.28e-01 | 1.00e+00 | 1.964 | 1 | 23 | 264 |
| GO:0043065 | positive regulation of apoptotic process | 2.31e-01 | 1.00e+00 | 1.943 | 1 | 7 | 268 |
| GO:0005743 | mitochondrial inner membrane | 2.38e-01 | 1.00e+00 | 1.895 | 1 | 6 | 277 |
| GO:0006200 | ATP catabolic process | 2.49e-01 | 1.00e+00 | 1.819 | 1 | 2 | 292 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.52e-01 | 1.00e+00 | 1.047 | 2 | 43 | 997 |
| GO:0008270 | zinc ion binding | 2.52e-01 | 1.00e+00 | 1.047 | 2 | 27 | 997 |
| GO:0005525 | GTP binding | 2.62e-01 | 1.00e+00 | 1.733 | 1 | 12 | 310 |
| GO:0003682 | chromatin binding | 2.75e-01 | 1.00e+00 | 1.656 | 1 | 19 | 327 |
| GO:0015031 | protein transport | 2.85e-01 | 1.00e+00 | 1.595 | 1 | 11 | 341 |
| GO:0043565 | sequence-specific DNA binding | 2.88e-01 | 1.00e+00 | 1.574 | 1 | 11 | 346 |
| GO:0000139 | Golgi membrane | 2.99e-01 | 1.00e+00 | 1.513 | 1 | 7 | 361 |
| GO:0005925 | focal adhesion | 3.02e-01 | 1.00e+00 | 1.493 | 1 | 9 | 366 |
| GO:0000278 | mitotic cell cycle | 3.20e-01 | 1.00e+00 | 1.398 | 1 | 15 | 391 |
| GO:0009986 | cell surface | 3.27e-01 | 1.00e+00 | 1.358 | 1 | 9 | 402 |
| GO:0044281 | small molecule metabolic process | 3.31e-01 | 1.00e+00 | 0.767 | 2 | 20 | 1211 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.34e-01 | 1.00e+00 | 1.319 | 1 | 24 | 413 |
| GO:0055114 | oxidation-reduction process | 3.39e-01 | 1.00e+00 | 1.294 | 1 | 10 | 420 |
| GO:0006351 | transcription, DNA-templated | 4.17e-01 | 1.00e+00 | 0.511 | 2 | 57 | 1446 |
| GO:0070062 | extracellular vesicular exosome | 4.19e-01 | 1.00e+00 | 0.365 | 3 | 57 | 2400 |
| GO:0006915 | apoptotic process | 4.23e-01 | 1.00e+00 | 0.892 | 1 | 12 | 555 |
| GO:0005783 | endoplasmic reticulum | 4.23e-01 | 1.00e+00 | 0.890 | 1 | 13 | 556 |
| GO:0005789 | endoplasmic reticulum membrane | 4.32e-01 | 1.00e+00 | 0.854 | 1 | 9 | 570 |
| GO:0005794 | Golgi apparatus | 4.54e-01 | 1.00e+00 | 0.756 | 1 | 9 | 610 |
| GO:0005886 | plasma membrane | 4.71e-01 | 1.00e+00 | 0.259 | 3 | 49 | 2582 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 5.01e-01 | 1.00e+00 | 0.560 | 1 | 39 | 699 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 5.45e-01 | 1.00e+00 | 0.385 | 1 | 41 | 789 |
| GO:0005615 | extracellular space | 6.18e-01 | 1.00e+00 | 0.106 | 1 | 20 | 957 |
| GO:0005739 | mitochondrion | 6.34e-01 | 1.00e+00 | 0.046 | 1 | 23 | 998 |
| GO:0005634 | nucleus | 6.92e-01 | 1.00e+00 | -0.146 | 4 | 159 | 4559 |
| GO:0046872 | metal ion binding | 7.36e-01 | 1.00e+00 | -0.343 | 1 | 29 | 1307 |
| GO:0005730 | nucleolus | 8.16e-01 | 1.00e+00 | -0.672 | 1 | 66 | 1641 |
| GO:0016021 | integral component of membrane | 8.74e-01 | 1.00e+00 | -0.944 | 1 | 17 | 1982 |