int-snw-7004
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 3.171 | 2.38e-19 | 4.75e-06 | 1.28e-04 |
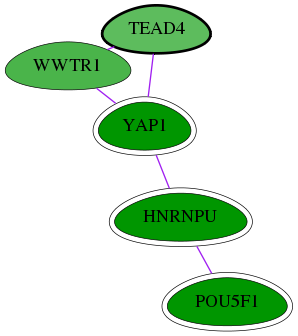
Genes (5)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| YAP1 | 10413 | 94 | -4.256 | 3.538 | 53 | Yes | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| HNRNPU | 3192 | 76 | -2.869 | 3.538 | 139 | Yes | - |
| WWTR1 | 25937 | 2 | -1.886 | 3.171 | 40 | - | - |
| [ TEAD4 ] | 7004 | 1 | -1.693 | 3.171 | 14 | - | - |
Interactions (5)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| TEAD4 | 7004 | WWTR1 | 25937 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid, BioGrid_Mouse, HPRD, VidalHuman_core, IntAct; int.Ravasi: -; int.HPRD: yeast 2-hybrid |
| YAP1 | 10413 | WWTR1 | 25937 | pp | -- | int.I2D: IntAct_Mouse |
| TEAD4 | 7004 | YAP1 | 10413 | pp | -- | int.I2D: HPRD, BioGrid_Mouse; int.Transfac: -; int.HPRD: in vitro, in vivo |
| HNRNPU | 3192 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| HNRNPU | 3192 | YAP1 | 10413 | pp | -- | int.I2D: HPRD; int.HPRD: in vitro, in vivo |
Related GO terms (77)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0035329 | hippo signaling | 5.19e-08 | 7.48e-04 | 8.379 | 3 | 5 | 26 |
| GO:0005654 | nucleoplasm | 2.36e-06 | 3.40e-02 | 3.737 | 5 | 64 | 1082 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 2.00e-05 | 2.89e-01 | 5.556 | 3 | 22 | 184 |
| GO:0010467 | gene expression | 2.21e-05 | 3.19e-01 | 4.108 | 4 | 45 | 669 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 3.47e-04 | 1.00e+00 | 11.494 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 3.47e-04 | 1.00e+00 | 11.494 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 3.47e-04 | 1.00e+00 | 11.494 | 1 | 1 | 1 |
| GO:0060390 | regulation of SMAD protein import into nucleus | 3.47e-04 | 1.00e+00 | 11.494 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 3.47e-04 | 1.00e+00 | 11.494 | 1 | 1 | 1 |
| GO:0001649 | osteoblast differentiation | 3.97e-04 | 1.00e+00 | 5.971 | 2 | 5 | 92 |
| GO:0009786 | regulation of asymmetric cell division | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 2 |
| GO:0060242 | contact inhibition | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 2 |
| GO:0060913 | cardiac cell fate determination | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 1.04e-03 | 1.00e+00 | 9.909 | 1 | 1 | 3 |
| GO:0044212 | transcription regulatory region DNA binding | 1.32e-03 | 1.00e+00 | 5.102 | 2 | 17 | 168 |
| GO:0005667 | transcription factor complex | 1.43e-03 | 1.00e+00 | 5.043 | 2 | 17 | 175 |
| GO:0003714 | transcription corepressor activity | 1.48e-03 | 1.00e+00 | 5.018 | 2 | 11 | 178 |
| GO:0001714 | endodermal cell fate specification | 1.73e-03 | 1.00e+00 | 9.172 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 1.73e-03 | 1.00e+00 | 9.172 | 1 | 2 | 5 |
| GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 1.73e-03 | 1.00e+00 | 9.172 | 1 | 1 | 5 |
| GO:0070937 | CRD-mediated mRNA stability complex | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 3 | 6 |
| GO:0017145 | stem cell division | 2.42e-03 | 1.00e+00 | 8.687 | 1 | 1 | 7 |
| GO:0003713 | transcription coactivator activity | 2.60e-03 | 1.00e+00 | 4.605 | 2 | 24 | 237 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.96e-03 | 1.00e+00 | 3.118 | 3 | 43 | 997 |
| GO:0032835 | glomerulus development | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 9 |
| GO:0035414 | negative regulation of catenin import into nucleus | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 9 |
| GO:0035413 | positive regulation of catenin import into nucleus | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 2 | 10 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 4 | 11 |
| GO:0035198 | miRNA binding | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 12 |
| GO:0071480 | cellular response to gamma radiation | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 13 |
| GO:0001824 | blastocyst development | 5.19e-03 | 1.00e+00 | 7.587 | 1 | 2 | 15 |
| GO:0003677 | DNA binding | 5.28e-03 | 1.00e+00 | 2.829 | 3 | 49 | 1218 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 2 | 22 |
| GO:0010718 | positive regulation of epithelial to mesenchymal transition | 8.64e-03 | 1.00e+00 | 6.850 | 1 | 4 | 25 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 2 | 32 |
| GO:0045599 | negative regulation of fat cell differentiation | 1.21e-02 | 1.00e+00 | 6.365 | 1 | 2 | 35 |
| GO:0001933 | negative regulation of protein phosphorylation | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 2 | 36 |
| GO:0005515 | protein binding | 1.27e-02 | 1.00e+00 | 1.260 | 5 | 198 | 6024 |
| GO:0060271 | cilium morphogenesis | 1.28e-02 | 1.00e+00 | 6.285 | 1 | 3 | 37 |
| GO:0035019 | somatic stem cell maintenance | 1.38e-02 | 1.00e+00 | 6.172 | 1 | 6 | 40 |
| GO:0009611 | response to wounding | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 5 | 52 |
| GO:0006396 | RNA processing | 2.03e-02 | 1.00e+00 | 5.612 | 1 | 5 | 59 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 2.13e-02 | 1.00e+00 | 3.045 | 2 | 39 | 699 |
| GO:0006469 | negative regulation of protein kinase activity | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 4 | 65 |
| GO:0010468 | regulation of gene expression | 2.34e-02 | 1.00e+00 | 5.407 | 1 | 2 | 68 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 2.54e-02 | 1.00e+00 | 5.285 | 1 | 4 | 74 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 2.68e-02 | 1.00e+00 | 2.870 | 2 | 41 | 789 |
| GO:0071013 | catalytic step 2 spliceosome | 2.68e-02 | 1.00e+00 | 5.209 | 1 | 7 | 78 |
| GO:0007517 | muscle organ development | 2.84e-02 | 1.00e+00 | 5.119 | 1 | 4 | 83 |
| GO:0009653 | anatomical structure morphogenesis | 2.84e-02 | 1.00e+00 | 5.119 | 1 | 2 | 83 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2.95e-02 | 1.00e+00 | 5.068 | 1 | 8 | 86 |
| GO:0005634 | nucleus | 3.73e-02 | 1.00e+00 | 1.340 | 4 | 159 | 4559 |
| GO:0030529 | ribonucleoprotein complex | 3.82e-02 | 1.00e+00 | 4.687 | 1 | 5 | 112 |
| GO:0005829 | cytosol | 3.93e-02 | 1.00e+00 | 1.794 | 3 | 86 | 2496 |
| GO:0001501 | skeletal system development | 4.06e-02 | 1.00e+00 | 4.599 | 1 | 2 | 119 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 4.36e-02 | 1.00e+00 | 4.494 | 1 | 11 | 128 |
| GO:0044822 | poly(A) RNA binding | 4.62e-02 | 1.00e+00 | 2.450 | 2 | 50 | 1056 |
| GO:0006974 | cellular response to DNA damage stimulus | 5.13e-02 | 1.00e+00 | 4.256 | 1 | 7 | 151 |
| GO:0000398 | mRNA splicing, via spliceosome | 5.56e-02 | 1.00e+00 | 4.137 | 1 | 15 | 164 |
| GO:0031625 | ubiquitin protein ligase binding | 6.02e-02 | 1.00e+00 | 4.018 | 1 | 5 | 178 |
| GO:0008380 | RNA splicing | 7.66e-02 | 1.00e+00 | 3.661 | 1 | 22 | 228 |
| GO:0008134 | transcription factor binding | 8.11e-02 | 1.00e+00 | 3.575 | 1 | 18 | 242 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 8.82e-02 | 1.00e+00 | 3.450 | 1 | 23 | 264 |
| GO:0008283 | cell proliferation | 1.09e-01 | 1.00e+00 | 3.137 | 1 | 14 | 328 |
| GO:0003723 | RNA binding | 1.13e-01 | 1.00e+00 | 3.076 | 1 | 18 | 342 |
| GO:0043565 | sequence-specific DNA binding | 1.14e-01 | 1.00e+00 | 3.060 | 1 | 11 | 346 |
| GO:0005737 | cytoplasm | 1.16e-01 | 1.00e+00 | 1.200 | 3 | 124 | 3767 |
| GO:0008284 | positive regulation of cell proliferation | 1.26e-01 | 1.00e+00 | 2.917 | 1 | 10 | 382 |
| GO:0009986 | cell surface | 1.32e-01 | 1.00e+00 | 2.843 | 1 | 9 | 402 |
| GO:0006366 | transcription from RNA polymerase II promoter | 1.37e-01 | 1.00e+00 | 2.783 | 1 | 30 | 419 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 1.83e-01 | 1.00e+00 | 2.339 | 1 | 37 | 570 |
| GO:0042803 | protein homodimerization activity | 1.90e-01 | 1.00e+00 | 2.277 | 1 | 19 | 595 |
| GO:0005524 | ATP binding | 3.76e-01 | 1.00e+00 | 1.152 | 1 | 31 | 1298 |
| GO:0006351 | transcription, DNA-templated | 4.10e-01 | 1.00e+00 | 0.996 | 1 | 57 | 1446 |
| GO:0005730 | nucleolus | 4.53e-01 | 1.00e+00 | 0.814 | 1 | 66 | 1641 |
| GO:0016020 | membrane | 4.62e-01 | 1.00e+00 | 0.779 | 1 | 46 | 1681 |