int-snw-6663
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 3.050 | 6.57e-18 | 2.89e-05 | 4.60e-04 |
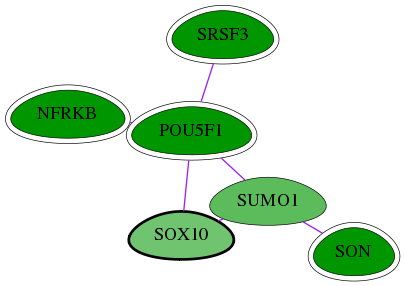
Genes (6)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| [ SOX10 ] | 6663 | 2 | -1.490 | 3.050 | 33 | - | - |
| SRSF3 | 6428 | 73 | -2.992 | 3.538 | 54 | Yes | - |
| NFRKB | 4798 | 48 | -3.129 | 3.108 | 23 | Yes | Yes |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| SON | 6651 | 9 | -3.844 | 3.108 | 9 | Yes | Yes |
| SUMO1 | 7341 | 7 | -1.696 | 3.050 | 439 | - | - |
Interactions (6)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| NFRKB | 4798 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| SOX10 | 6663 | SUMO1 | 7341 | pp | -- | int.I2D: BCI, HPRD, MINT; int.Mint: MI:0566(sumoylation reaction); int.HPRD: in vivo |
| POU5F1 | 5460 | SOX10 | 6663 | pp | -- | int.I2D: HPRD, BCI; int.Ravasi: -; int.HPRD: in vitro |
| POU5F1 | 5460 | SRSF3 | 6428 | pp | -- | int.I2D: BioGrid |
| SON | 6651 | SUMO1 | 7341 | pp | -- | int.I2D: BioGrid |
| POU5F1 | 5460 | SUMO1 | 7341 | pp | -- | int.I2D: BioGrid, BioGrid_Mouse |
Related GO terms (101)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0006366 | transcription from RNA polymerase II promoter | 1.01e-05 | 1.45e-01 | 4.520 | 4 | 30 | 419 |
| GO:0016607 | nuclear speck | 3.25e-05 | 4.68e-01 | 5.390 | 3 | 12 | 172 |
| GO:0008134 | transcription factor binding | 8.99e-05 | 1.00e+00 | 4.897 | 3 | 18 | 242 |
| GO:0044822 | poly(A) RNA binding | 3.80e-04 | 1.00e+00 | 3.187 | 4 | 50 | 1056 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0009653 | anatomical structure morphogenesis | 4.83e-04 | 1.00e+00 | 5.856 | 2 | 2 | 83 |
| GO:0090204 | protein localization to nuclear pore | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0031315 | extrinsic component of mitochondrial outer membrane | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0014015 | positive regulation of gliogenesis | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0060913 | cardiac cell fate determination | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0001650 | fibrillar center | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0030578 | PML body organization | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 4 |
| GO:0001714 | endodermal cell fate specification | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 5 |
| GO:0031625 | ubiquitin protein ligase binding | 2.20e-03 | 1.00e+00 | 4.755 | 2 | 5 | 178 |
| GO:0050733 | RS domain binding | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 6 |
| GO:0008380 | RNA splicing | 3.58e-03 | 1.00e+00 | 4.398 | 2 | 22 | 228 |
| GO:0035413 | positive regulation of catenin import into nucleus | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 2 | 10 |
| GO:0048484 | enteric nervous system development | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 10 |
| GO:0019789 | SUMO ligase activity | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 4 | 11 |
| GO:0006281 | DNA repair | 4.66e-03 | 1.00e+00 | 4.203 | 2 | 14 | 261 |
| GO:0035198 | miRNA binding | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 12 |
| GO:0005515 | protein binding | 5.30e-03 | 1.00e+00 | 1.260 | 6 | 198 | 6024 |
| GO:0031334 | positive regulation of protein complex assembly | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 2 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 2 | 13 |
| GO:0031011 | Ino80 complex | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 6 | 14 |
| GO:0048546 | digestive tract morphogenesis | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 14 |
| GO:0001824 | blastocyst development | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 2 | 15 |
| GO:0043274 | phospholipase binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0060334 | regulation of interferon-gamma-mediated signaling pathway | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0001190 | RNA polymerase II transcription factor binding transcription factor activity involved in positive regulation of transcription | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 2 | 16 |
| GO:0005654 | nucleoplasm | 7.09e-03 | 1.00e+00 | 2.737 | 3 | 64 | 1082 |
| GO:0002052 | positive regulation of neuroblast proliferation | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 1 | 18 |
| GO:0030318 | melanocyte differentiation | 7.88e-03 | 1.00e+00 | 6.983 | 1 | 3 | 19 |
| GO:0003723 | RNA binding | 7.89e-03 | 1.00e+00 | 3.813 | 2 | 18 | 342 |
| GO:0043484 | regulation of RNA splicing | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 22 |
| GO:0048709 | oligodendrocyte differentiation | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 2 | 23 |
| GO:0003677 | DNA binding | 9.89e-03 | 1.00e+00 | 2.566 | 3 | 49 | 1218 |
| GO:0007422 | peripheral nervous system development | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 25 |
| GO:0016925 | protein sumoylation | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 25 |
| GO:0048589 | developmental growth | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 25 |
| GO:0043392 | negative regulation of DNA binding | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 4 | 26 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 1.14e-02 | 1.00e+00 | 3.541 | 2 | 24 | 413 |
| GO:0001755 | neural crest cell migration | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 1 | 28 |
| GO:0043066 | negative regulation of apoptotic process | 1.20e-02 | 1.00e+00 | 3.503 | 2 | 16 | 424 |
| GO:0005634 | nucleus | 1.39e-02 | 1.00e+00 | 1.399 | 5 | 159 | 4559 |
| GO:0032880 | regulation of protein localization | 1.53e-02 | 1.00e+00 | 6.022 | 1 | 2 | 37 |
| GO:0048469 | cell maturation | 1.57e-02 | 1.00e+00 | 5.983 | 1 | 1 | 38 |
| GO:0035019 | somatic stem cell maintenance | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 6 | 40 |
| GO:0031124 | mRNA 3'-end processing | 1.73e-02 | 1.00e+00 | 5.839 | 1 | 2 | 42 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 3 | 44 |
| GO:0006369 | termination of RNA polymerase II transcription | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 2 | 45 |
| GO:0000910 | cytokinesis | 2.10e-02 | 1.00e+00 | 5.559 | 1 | 2 | 51 |
| GO:0009611 | response to wounding | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 5 | 52 |
| GO:0000226 | microtubule cytoskeleton organization | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 2 | 52 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 2.19e-02 | 1.00e+00 | 5.503 | 1 | 5 | 53 |
| GO:0005730 | nucleolus | 2.26e-02 | 1.00e+00 | 2.136 | 3 | 66 | 1641 |
| GO:0005643 | nuclear pore | 2.35e-02 | 1.00e+00 | 5.398 | 1 | 11 | 57 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 2.43e-02 | 1.00e+00 | 5.348 | 1 | 6 | 59 |
| GO:0003705 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity | 2.51e-02 | 1.00e+00 | 5.300 | 1 | 4 | 61 |
| GO:0006406 | mRNA export from nucleus | 2.51e-02 | 1.00e+00 | 5.300 | 1 | 4 | 61 |
| GO:0060333 | interferon-gamma-mediated signaling pathway | 2.55e-02 | 1.00e+00 | 5.277 | 1 | 3 | 62 |
| GO:0006310 | DNA recombination | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 7 | 65 |
| GO:0010468 | regulation of gene expression | 2.80e-02 | 1.00e+00 | 5.144 | 1 | 2 | 68 |
| GO:0060021 | palate development | 2.96e-02 | 1.00e+00 | 5.061 | 1 | 1 | 72 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 4 | 74 |
| GO:0002020 | protease binding | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 6 | 74 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 3.53e-02 | 1.00e+00 | 4.805 | 1 | 8 | 86 |
| GO:0016605 | PML body | 3.61e-02 | 1.00e+00 | 4.772 | 1 | 3 | 88 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 3.87e-02 | 1.00e+00 | 2.607 | 2 | 41 | 789 |
| GO:0051726 | regulation of cell cycle | 3.97e-02 | 1.00e+00 | 4.631 | 1 | 5 | 97 |
| GO:0003676 | nucleic acid binding | 4.37e-02 | 1.00e+00 | 4.490 | 1 | 2 | 107 |
| GO:0045202 | synapse | 5.53e-02 | 1.00e+00 | 4.144 | 1 | 5 | 136 |
| GO:0006355 | regulation of transcription, DNA-templated | 5.94e-02 | 1.00e+00 | 2.270 | 2 | 43 | 997 |
| GO:0006397 | mRNA processing | 6.48e-02 | 1.00e+00 | 3.909 | 1 | 10 | 160 |
| GO:0043687 | post-translational protein modification | 6.55e-02 | 1.00e+00 | 3.891 | 1 | 5 | 162 |
| GO:0000398 | mRNA splicing, via spliceosome | 6.63e-02 | 1.00e+00 | 3.874 | 1 | 15 | 164 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 6.67e-02 | 1.00e+00 | 3.865 | 1 | 12 | 165 |
| GO:0031965 | nuclear membrane | 6.79e-02 | 1.00e+00 | 3.839 | 1 | 10 | 168 |
| GO:0044212 | transcription regulatory region DNA binding | 6.79e-02 | 1.00e+00 | 3.839 | 1 | 17 | 168 |
| GO:0005667 | transcription factor complex | 7.06e-02 | 1.00e+00 | 3.780 | 1 | 17 | 175 |
| GO:0001701 | in utero embryonic development | 8.08e-02 | 1.00e+00 | 3.580 | 1 | 8 | 201 |
| GO:0019221 | cytokine-mediated signaling pathway | 8.85e-02 | 1.00e+00 | 3.443 | 1 | 8 | 221 |
| GO:0030425 | dendrite | 9.20e-02 | 1.00e+00 | 3.386 | 1 | 6 | 230 |
| GO:0003713 | transcription coactivator activity | 9.46e-02 | 1.00e+00 | 3.342 | 1 | 24 | 237 |
| GO:0000166 | nucleotide binding | 1.03e-01 | 1.00e+00 | 3.214 | 1 | 13 | 259 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 1.05e-01 | 1.00e+00 | 3.187 | 1 | 23 | 264 |
| GO:0006954 | inflammatory response | 1.10e-01 | 1.00e+00 | 3.117 | 1 | 6 | 277 |
| GO:0003682 | chromatin binding | 1.29e-01 | 1.00e+00 | 2.878 | 1 | 19 | 327 |
| GO:0043565 | sequence-specific DNA binding | 1.36e-01 | 1.00e+00 | 2.796 | 1 | 11 | 346 |
| GO:0044267 | cellular protein metabolic process | 1.82e-01 | 1.00e+00 | 2.342 | 1 | 20 | 474 |
| GO:0042802 | identical protein binding | 1.85e-01 | 1.00e+00 | 2.312 | 1 | 19 | 484 |
| GO:0005737 | cytoplasm | 1.87e-01 | 1.00e+00 | 0.937 | 3 | 124 | 3767 |
| GO:0010467 | gene expression | 2.48e-01 | 1.00e+00 | 1.845 | 1 | 45 | 669 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 2.58e-01 | 1.00e+00 | 1.782 | 1 | 39 | 699 |
| GO:0005829 | cytosol | 6.80e-01 | 1.00e+00 | -0.054 | 1 | 86 | 2496 |