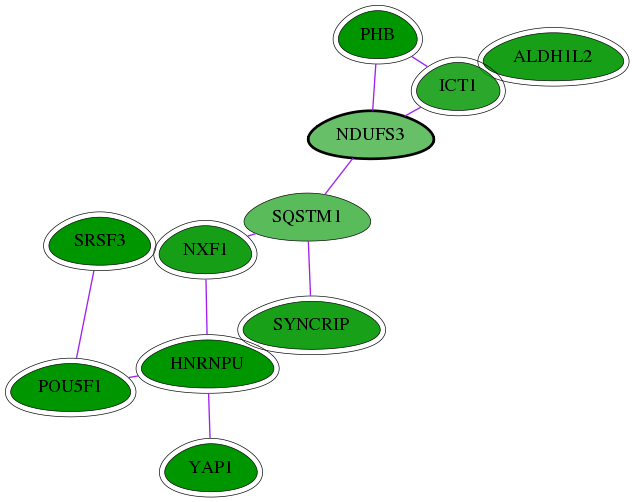
| GO:0005654 | nucleoplasm | 1.32e-07 | 1.90e-03 | 3.277 | 8 | 64 | 1082 |
| GO:0070934 | CRD-mediated mRNA stabilization | 5.28e-06 | 7.62e-02 | 9.035 | 2 | 2 | 5 |
| GO:0070937 | CRD-mediated mRNA stability complex | 7.92e-06 | 1.14e-01 | 8.772 | 2 | 3 | 6 |
| GO:0001649 | osteoblast differentiation | 3.99e-05 | 5.76e-01 | 5.418 | 3 | 5 | 92 |
| GO:0000398 | mRNA splicing, via spliceosome | 2.23e-04 | 1.00e+00 | 4.584 | 3 | 15 | 164 |
| GO:0044212 | transcription regulatory region DNA binding | 2.39e-04 | 1.00e+00 | 4.549 | 3 | 17 | 168 |
| GO:0008380 | RNA splicing | 5.86e-04 | 1.00e+00 | 4.109 | 3 | 22 | 228 |
| GO:0044822 | poly(A) RNA binding | 6.63e-04 | 1.00e+00 | 2.634 | 5 | 50 | 1056 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0070126 | mitochondrial translational termination | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0016150 | translation release factor activity, codon nonspecific | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0000166 | nucleotide binding | 8.49e-04 | 1.00e+00 | 3.925 | 3 | 13 | 259 |
| GO:0006396 | RNA processing | 8.84e-04 | 1.00e+00 | 5.474 | 2 | 5 | 59 |
| GO:0006406 | mRNA export from nucleus | 9.44e-04 | 1.00e+00 | 5.426 | 2 | 4 | 61 |
| GO:0010467 | gene expression | 1.17e-03 | 1.00e+00 | 2.971 | 4 | 45 | 669 |
| GO:0060242 | contact inhibition | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0016155 | formyltetrahydrofolate dehydrogenase activity | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0009258 | 10-formyltetrahydrofolate catabolic process | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0016742 | hydroxymethyl-, formyl- and related transferase activity | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0071013 | catalytic step 2 spliceosome | 1.54e-03 | 1.00e+00 | 5.071 | 2 | 7 | 78 |
| GO:0003723 | RNA binding | 1.89e-03 | 1.00e+00 | 3.524 | 3 | 18 | 342 |
| GO:0004045 | aminoacyl-tRNA hydrolase activity | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0030308 | negative regulation of cell growth | 3.03e-03 | 1.00e+00 | 4.575 | 2 | 6 | 110 |
| GO:0097452 | GAIT complex | 3.05e-03 | 1.00e+00 | 8.357 | 1 | 1 | 4 |
| GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 3.05e-03 | 1.00e+00 | 8.357 | 1 | 1 | 4 |
| GO:0030529 | ribonucleoprotein complex | 3.14e-03 | 1.00e+00 | 4.549 | 2 | 5 | 112 |
| GO:0001714 | endodermal cell fate specification | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 1 | 5 |
| GO:0050847 | progesterone receptor signaling pathway | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 2 | 6 |
| GO:0003954 | NADH dehydrogenase activity | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 6 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 6 |
| GO:0016236 | macroautophagy | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 6 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 6 |
| GO:0005739 | mitochondrion | 5.07e-03 | 1.00e+00 | 2.394 | 4 | 23 | 998 |
| GO:0071354 | cellular response to interleukin-6 | 5.33e-03 | 1.00e+00 | 7.549 | 1 | 2 | 7 |
| GO:0046578 | regulation of Ras protein signal transduction | 5.33e-03 | 1.00e+00 | 7.549 | 1 | 1 | 7 |
| GO:0016973 | poly(A)+ mRNA export from nucleus | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 1 | 8 |
| GO:0042405 | nuclear inclusion body | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 2 | 9 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 1 | 9 |
| GO:0016607 | nuclear speck | 7.25e-03 | 1.00e+00 | 3.930 | 2 | 12 | 172 |
| GO:0035413 | positive regulation of catenin import into nucleus | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 2 | 10 |
| GO:0009058 | biosynthetic process | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 1 | 10 |
| GO:0005515 | protein binding | 8.30e-03 | 1.00e+00 | 0.970 | 9 | 198 | 6024 |
| GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 8.36e-03 | 1.00e+00 | 6.897 | 1 | 1 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 8.36e-03 | 1.00e+00 | 6.897 | 1 | 4 | 11 |
| GO:0070530 | K63-linked polyubiquitin binding | 8.36e-03 | 1.00e+00 | 6.897 | 1 | 2 | 11 |
| GO:0016239 | positive regulation of macroautophagy | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 12 |
| GO:0008143 | poly(A) binding | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 2 | 12 |
| GO:0035198 | miRNA binding | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 12 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 3 | 13 |
| GO:0071480 | cellular response to gamma radiation | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 2 | 13 |
| GO:0000407 | pre-autophagosomal structure | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 2 | 13 |
| GO:0000346 | transcription export complex | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 13 |
| GO:0005762 | mitochondrial large ribosomal subunit | 1.06e-02 | 1.00e+00 | 6.549 | 1 | 1 | 14 |
| GO:0005487 | nucleocytoplasmic transporter activity | 1.06e-02 | 1.00e+00 | 6.549 | 1 | 3 | 14 |
| GO:0001824 | blastocyst development | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 2 | 15 |
| GO:0043274 | phospholipase binding | 1.21e-02 | 1.00e+00 | 6.357 | 1 | 1 | 16 |
| GO:0016234 | inclusion body | 1.44e-02 | 1.00e+00 | 6.109 | 1 | 2 | 19 |
| GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 1.59e-02 | 1.00e+00 | 5.964 | 1 | 1 | 21 |
| GO:0016575 | histone deacetylation | 1.59e-02 | 1.00e+00 | 5.964 | 1 | 3 | 21 |
| GO:0016235 | aggresome | 1.59e-02 | 1.00e+00 | 5.964 | 1 | 1 | 21 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.67e-02 | 1.00e+00 | 5.897 | 1 | 2 | 22 |
| GO:0071346 | cellular response to interferon-gamma | 1.74e-02 | 1.00e+00 | 5.833 | 1 | 1 | 23 |
| GO:0006730 | one-carbon metabolic process | 1.74e-02 | 1.00e+00 | 5.833 | 1 | 1 | 23 |
| GO:0005743 | mitochondrial inner membrane | 1.80e-02 | 1.00e+00 | 3.243 | 2 | 6 | 277 |
| GO:0035329 | hippo signaling | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 5 | 26 |
| GO:0071897 | DNA biosynthetic process | 2.12e-02 | 1.00e+00 | 5.549 | 1 | 1 | 28 |
| GO:0042169 | SH2 domain binding | 2.27e-02 | 1.00e+00 | 5.450 | 1 | 2 | 30 |
| GO:0072593 | reactive oxygen species metabolic process | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 2 | 31 |
| GO:0002376 | immune system process | 2.41e-02 | 1.00e+00 | 5.357 | 1 | 3 | 32 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 2.41e-02 | 1.00e+00 | 5.357 | 1 | 2 | 32 |
| GO:0005776 | autophagic vacuole | 2.41e-02 | 1.00e+00 | 5.357 | 1 | 1 | 32 |
| GO:0030971 | receptor tyrosine kinase binding | 2.49e-02 | 1.00e+00 | 5.312 | 1 | 2 | 33 |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | 2.71e-02 | 1.00e+00 | 5.187 | 1 | 1 | 36 |
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 2.71e-02 | 1.00e+00 | 5.187 | 1 | 1 | 36 |
| GO:0005747 | mitochondrial respiratory chain complex I | 2.94e-02 | 1.00e+00 | 5.071 | 1 | 1 | 39 |
| GO:0035019 | somatic stem cell maintenance | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 6 | 40 |
| GO:0017148 | negative regulation of translation | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 2 | 40 |
| GO:0008168 | methyltransferase activity | 3.08e-02 | 1.00e+00 | 4.999 | 1 | 2 | 41 |
| GO:0031124 | mRNA 3'-end processing | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 2 | 42 |
| GO:0005080 | protein kinase C binding | 3.23e-02 | 1.00e+00 | 4.930 | 1 | 1 | 43 |
| GO:0021762 | substantia nigra development | 3.31e-02 | 1.00e+00 | 4.897 | 1 | 4 | 44 |
| GO:0043130 | ubiquitin binding | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 2 | 45 |
| GO:0006369 | termination of RNA polymerase II transcription | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 2 | 45 |
| GO:0006950 | response to stress | 3.45e-02 | 1.00e+00 | 4.833 | 1 | 2 | 46 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.57e-02 | 1.00e+00 | 1.980 | 3 | 43 | 997 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.89e-02 | 1.00e+00 | 2.646 | 2 | 30 | 419 |
| GO:0009611 | response to wounding | 3.90e-02 | 1.00e+00 | 4.656 | 1 | 5 | 52 |
| GO:0000932 | cytoplasmic mRNA processing body | 4.04e-02 | 1.00e+00 | 4.602 | 1 | 4 | 54 |
| GO:0031966 | mitochondrial membrane | 4.12e-02 | 1.00e+00 | 4.575 | 1 | 1 | 55 |
| GO:0008104 | protein localization | 4.19e-02 | 1.00e+00 | 4.549 | 1 | 1 | 56 |
| GO:0051291 | protein heterooligomerization | 4.19e-02 | 1.00e+00 | 4.549 | 1 | 4 | 56 |
| GO:0032259 | methylation | 4.26e-02 | 1.00e+00 | 4.524 | 1 | 1 | 57 |
| GO:0005643 | nuclear pore | 4.26e-02 | 1.00e+00 | 4.524 | 1 | 11 | 57 |
| GO:0016197 | endosomal transport | 4.26e-02 | 1.00e+00 | 4.524 | 1 | 2 | 57 |
| GO:0009055 | electron carrier activity | 4.85e-02 | 1.00e+00 | 4.334 | 1 | 2 | 65 |
| GO:0010468 | regulation of gene expression | 5.07e-02 | 1.00e+00 | 4.269 | 1 | 2 | 68 |
| GO:0042826 | histone deacetylase binding | 5.36e-02 | 1.00e+00 | 4.187 | 1 | 4 | 72 |
| GO:0003729 | mRNA binding | 5.36e-02 | 1.00e+00 | 4.187 | 1 | 4 | 72 |
| GO:0006914 | autophagy | 5.50e-02 | 1.00e+00 | 4.147 | 1 | 3 | 74 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 5.50e-02 | 1.00e+00 | 4.147 | 1 | 4 | 74 |
| GO:0016032 | viral process | 6.03e-02 | 1.00e+00 | 2.296 | 2 | 26 | 534 |
| GO:0009653 | anatomical structure morphogenesis | 6.15e-02 | 1.00e+00 | 3.982 | 1 | 2 | 83 |
| GO:0022904 | respiratory electron transport chain | 6.30e-02 | 1.00e+00 | 3.947 | 1 | 1 | 85 |
| GO:0005783 | endoplasmic reticulum | 6.48e-02 | 1.00e+00 | 2.238 | 2 | 13 | 556 |
| GO:0016605 | PML body | 6.51e-02 | 1.00e+00 | 3.897 | 1 | 3 | 88 |
| GO:0005770 | late endosome | 6.87e-02 | 1.00e+00 | 3.817 | 1 | 2 | 93 |
| GO:0031410 | cytoplasmic vesicle | 7.58e-02 | 1.00e+00 | 3.670 | 1 | 4 | 103 |
| GO:0001934 | positive regulation of protein phosphorylation | 7.65e-02 | 1.00e+00 | 3.656 | 1 | 5 | 104 |
| GO:0097190 | apoptotic signaling pathway | 8.29e-02 | 1.00e+00 | 3.536 | 1 | 3 | 113 |
| GO:0044237 | cellular metabolic process | 8.57e-02 | 1.00e+00 | 3.486 | 1 | 2 | 117 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 9.20e-02 | 1.00e+00 | 3.379 | 1 | 5 | 126 |
| GO:0005634 | nucleus | 9.78e-02 | 1.00e+00 | 0.787 | 6 | 159 | 4559 |
| GO:0005829 | cytosol | 1.07e-01 | 1.00e+00 | 1.071 | 4 | 86 | 2496 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.09e-01 | 1.00e+00 | 3.118 | 1 | 7 | 151 |
| GO:0042981 | regulation of apoptotic process | 1.09e-01 | 1.00e+00 | 3.128 | 1 | 4 | 150 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 1.19e-01 | 1.00e+00 | 1.733 | 2 | 41 | 789 |
| GO:0005764 | lysosome | 1.24e-01 | 1.00e+00 | 2.930 | 1 | 2 | 172 |
| GO:0005667 | transcription factor complex | 1.26e-01 | 1.00e+00 | 2.905 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.28e-01 | 1.00e+00 | 2.881 | 1 | 5 | 178 |
| GO:0003714 | transcription corepressor activity | 1.28e-01 | 1.00e+00 | 2.881 | 1 | 11 | 178 |
| GO:0016020 | membrane | 1.28e-01 | 1.00e+00 | 1.227 | 3 | 46 | 1681 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.32e-01 | 1.00e+00 | 2.833 | 1 | 22 | 184 |
| GO:0005737 | cytoplasm | 1.33e-01 | 1.00e+00 | 0.799 | 5 | 124 | 3767 |
| GO:0003713 | transcription coactivator activity | 1.67e-01 | 1.00e+00 | 2.468 | 1 | 24 | 237 |
| GO:0008134 | transcription factor binding | 1.70e-01 | 1.00e+00 | 2.438 | 1 | 18 | 242 |
| GO:0043065 | positive regulation of apoptotic process | 1.86e-01 | 1.00e+00 | 2.291 | 1 | 7 | 268 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 1.87e-01 | 1.00e+00 | 2.285 | 1 | 7 | 269 |
| GO:0019899 | enzyme binding | 1.92e-01 | 1.00e+00 | 2.243 | 1 | 9 | 277 |
| GO:0030154 | cell differentiation | 2.07e-01 | 1.00e+00 | 2.123 | 1 | 5 | 301 |
| GO:0035556 | intracellular signal transduction | 2.08e-01 | 1.00e+00 | 2.113 | 1 | 9 | 303 |
| GO:0004674 | protein serine/threonine kinase activity | 2.11e-01 | 1.00e+00 | 2.090 | 1 | 6 | 308 |
| GO:0019901 | protein kinase binding | 2.17e-01 | 1.00e+00 | 2.048 | 1 | 18 | 317 |
| GO:0008283 | cell proliferation | 2.24e-01 | 1.00e+00 | 1.999 | 1 | 14 | 328 |
| GO:0043565 | sequence-specific DNA binding | 2.34e-01 | 1.00e+00 | 1.922 | 1 | 11 | 346 |
| GO:0003677 | DNA binding | 2.37e-01 | 1.00e+00 | 1.106 | 2 | 49 | 1218 |
| GO:0008285 | negative regulation of cell proliferation | 2.39e-01 | 1.00e+00 | 1.893 | 1 | 10 | 353 |
| GO:0009986 | cell surface | 2.67e-01 | 1.00e+00 | 1.706 | 1 | 9 | 402 |
| GO:0070062 | extracellular vesicular exosome | 2.72e-01 | 1.00e+00 | 0.713 | 3 | 57 | 2400 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.74e-01 | 1.00e+00 | 1.667 | 1 | 24 | 413 |
| GO:0055114 | oxidation-reduction process | 2.78e-01 | 1.00e+00 | 1.642 | 1 | 10 | 420 |
| GO:0043066 | negative regulation of apoptotic process | 2.80e-01 | 1.00e+00 | 1.629 | 1 | 16 | 424 |
| GO:0006468 | protein phosphorylation | 3.00e-01 | 1.00e+00 | 1.511 | 1 | 10 | 460 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 3.10e-01 | 1.00e+00 | 1.456 | 1 | 24 | 478 |
| GO:0042802 | identical protein binding | 3.13e-01 | 1.00e+00 | 1.438 | 1 | 19 | 484 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.58e-01 | 1.00e+00 | 1.202 | 1 | 37 | 570 |
| GO:0042803 | protein homodimerization activity | 3.71e-01 | 1.00e+00 | 1.140 | 1 | 19 | 595 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 4.21e-01 | 1.00e+00 | 0.908 | 1 | 39 | 699 |
| GO:0005887 | integral component of plasma membrane | 5.03e-01 | 1.00e+00 | 0.562 | 1 | 10 | 888 |
| GO:0007165 | signal transduction | 5.11e-01 | 1.00e+00 | 0.532 | 1 | 13 | 907 |
| GO:0008270 | zinc ion binding | 5.45e-01 | 1.00e+00 | 0.395 | 1 | 27 | 997 |
| GO:0044281 | small molecule metabolic process | 6.19e-01 | 1.00e+00 | 0.115 | 1 | 20 | 1211 |
| GO:0005524 | ATP binding | 6.46e-01 | 1.00e+00 | 0.015 | 1 | 31 | 1298 |