int-snw-29915
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.853 | 1.07e-15 | 3.90e-04 | 2.91e-03 |
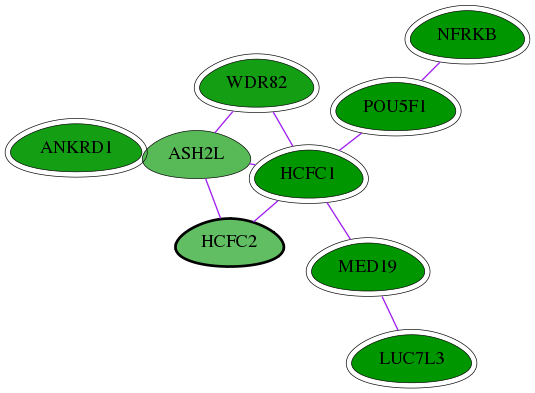
Genes (9)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| HCFC1 | 3054 | 40 | -2.925 | 3.449 | 71 | Yes | Yes |
| MED19 | 219541 | 30 | -2.657 | 3.449 | 115 | Yes | - |
| LUC7L3 | 51747 | 33 | -3.523 | 3.449 | 18 | Yes | - |
| ASH2L | 9070 | 1 | -1.756 | 2.853 | 65 | - | - |
| WDR82 | 80335 | 7 | -2.455 | 3.129 | 64 | Yes | - |
| NFRKB | 4798 | 48 | -3.129 | 3.108 | 23 | Yes | Yes |
| ANKRD1 | 27063 | 1 | -2.448 | 2.853 | 15 | Yes | Yes |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| [ HCFC2 ] | 29915 | 1 | -1.637 | 2.853 | 13 | - | - |
Interactions (10)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| ASH2L | 9070 | ANKRD1 | 27063 | pp | -- | int.Intact: MI:0915(physical association) |
| ASH2L | 9070 | WDR82 | 80335 | pp | -- | int.I2D: BioGrid |
| ASH2L | 9070 | HCFC2 | 29915 | pp | -- | int.I2D: BioGrid |
| NFRKB | 4798 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| LUC7L3 | 51747 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| HCFC1 | 3054 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| HCFC1 | 3054 | ASH2L | 9070 | pp | -- | int.Intact: MI:0915(physical association), MI:0914(association); int.I2D: BCI, IntAct, BioGrid; int.Ravasi: - |
| HCFC1 | 3054 | MED19 | 219541 | pp | -- | int.Intact: MI:0914(association); int.I2D: IntAct |
| HCFC1 | 3054 | HCFC2 | 29915 | pp | -- | int.I2D: BioGrid |
| HCFC1 | 3054 | WDR82 | 80335 | pp | -- | int.I2D: BioGrid |
Related GO terms (116)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0048188 | Set1C/COMPASS complex | 1.41e-08 | 2.03e-04 | 9.061 | 3 | 4 | 9 |
| GO:0006366 | transcription from RNA polymerase II promoter | 2.31e-06 | 3.33e-02 | 4.257 | 5 | 30 | 419 |
| GO:0042800 | histone methyltransferase activity (H3-K4 specific) | 2.69e-05 | 3.88e-01 | 7.946 | 2 | 3 | 13 |
| GO:0051568 | histone H3-K4 methylation | 4.68e-05 | 6.76e-01 | 7.559 | 2 | 3 | 17 |
| GO:0035097 | histone methyltransferase complex | 7.22e-05 | 1.00e+00 | 7.254 | 2 | 3 | 21 |
| GO:0003677 | DNA binding | 4.02e-04 | 1.00e+00 | 2.718 | 5 | 49 | 1218 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 6.24e-04 | 1.00e+00 | 10.646 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 6.24e-04 | 1.00e+00 | 10.646 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 6.24e-04 | 1.00e+00 | 10.646 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 6.24e-04 | 1.00e+00 | 10.646 | 1 | 1 | 1 |
| GO:0005634 | nucleus | 6.43e-04 | 1.00e+00 | 1.492 | 8 | 159 | 4559 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 8.97e-04 | 1.00e+00 | 3.022 | 4 | 41 | 789 |
| GO:0019046 | release from viral latency | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 2 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 2 |
| GO:0060913 | cardiac cell fate determination | 1.87e-03 | 1.00e+00 | 9.061 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 1.87e-03 | 1.00e+00 | 9.061 | 1 | 1 | 3 |
| GO:0035994 | response to muscle stretch | 1.87e-03 | 1.00e+00 | 9.061 | 1 | 1 | 3 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.16e-03 | 1.00e+00 | 2.685 | 4 | 43 | 997 |
| GO:0070461 | SAGA-type complex | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 2 | 5 |
| GO:0001714 | endodermal cell fate specification | 3.12e-03 | 1.00e+00 | 8.324 | 1 | 1 | 5 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 4.31e-03 | 1.00e+00 | 3.076 | 3 | 37 | 570 |
| GO:0072357 | PTW/PP1 phosphatase complex | 4.36e-03 | 1.00e+00 | 7.839 | 1 | 2 | 7 |
| GO:0044212 | transcription regulatory region DNA binding | 4.60e-03 | 1.00e+00 | 4.254 | 2 | 17 | 168 |
| GO:2000279 | negative regulation of DNA biosynthetic process | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 8 |
| GO:0070688 | MLL5-L complex | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 8 |
| GO:0043254 | regulation of protein complex assembly | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 8 |
| GO:0005667 | transcription factor complex | 4.98e-03 | 1.00e+00 | 4.195 | 2 | 17 | 175 |
| GO:0043995 | histone acetyltransferase activity (H4-K5 specific) | 5.60e-03 | 1.00e+00 | 7.476 | 1 | 5 | 9 |
| GO:0046972 | histone acetyltransferase activity (H4-K16 specific) | 5.60e-03 | 1.00e+00 | 7.476 | 1 | 5 | 9 |
| GO:0043996 | histone acetyltransferase activity (H4-K8 specific) | 5.60e-03 | 1.00e+00 | 7.476 | 1 | 5 | 9 |
| GO:0035413 | positive regulation of catenin import into nucleus | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 2 | 10 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 4 | 11 |
| GO:0055008 | cardiac muscle tissue morphogenesis | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 1 | 12 |
| GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 2 | 12 |
| GO:0035198 | miRNA binding | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 1 | 12 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 8.09e-03 | 1.00e+00 | 6.946 | 1 | 2 | 13 |
| GO:0005685 | U1 snRNP | 8.70e-03 | 1.00e+00 | 6.839 | 1 | 1 | 14 |
| GO:0031011 | Ino80 complex | 8.70e-03 | 1.00e+00 | 6.839 | 1 | 6 | 14 |
| GO:0031432 | titin binding | 8.70e-03 | 1.00e+00 | 6.839 | 1 | 1 | 14 |
| GO:0003713 | transcription coactivator activity | 8.97e-03 | 1.00e+00 | 3.757 | 2 | 24 | 237 |
| GO:0043981 | histone H4-K5 acetylation | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 5 | 15 |
| GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 3 | 15 |
| GO:0006376 | mRNA splice site selection | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 1 | 15 |
| GO:0001824 | blastocyst development | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 2 | 15 |
| GO:0043982 | histone H4-K8 acetylation | 9.32e-03 | 1.00e+00 | 6.739 | 1 | 5 | 15 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 1.10e-02 | 1.00e+00 | 3.602 | 2 | 23 | 264 |
| GO:0031674 | I band | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 1 | 18 |
| GO:0043984 | histone H4-K16 acetylation | 1.18e-02 | 1.00e+00 | 6.398 | 1 | 5 | 19 |
| GO:0000123 | histone acetyltransferase complex | 1.30e-02 | 1.00e+00 | 6.254 | 1 | 6 | 21 |
| GO:0070412 | R-SMAD binding | 1.30e-02 | 1.00e+00 | 6.254 | 1 | 1 | 21 |
| GO:0005730 | nucleolus | 1.31e-02 | 1.00e+00 | 1.966 | 4 | 66 | 1641 |
| GO:0045787 | positive regulation of cell cycle | 1.36e-02 | 1.00e+00 | 6.187 | 1 | 2 | 22 |
| GO:0001205 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 1.36e-02 | 1.00e+00 | 6.187 | 1 | 4 | 22 |
| GO:0001105 | RNA polymerase II transcription coactivator activity | 1.49e-02 | 1.00e+00 | 6.061 | 1 | 2 | 24 |
| GO:0045214 | sarcomere organization | 1.61e-02 | 1.00e+00 | 5.946 | 1 | 2 | 26 |
| GO:0003682 | chromatin binding | 1.66e-02 | 1.00e+00 | 3.293 | 2 | 19 | 327 |
| GO:0071339 | MLL1 complex | 1.67e-02 | 1.00e+00 | 5.891 | 1 | 6 | 27 |
| GO:0050714 | positive regulation of protein secretion | 1.73e-02 | 1.00e+00 | 5.839 | 1 | 3 | 28 |
| GO:0071347 | cellular response to interleukin-1 | 1.92e-02 | 1.00e+00 | 5.692 | 1 | 2 | 31 |
| GO:0001085 | RNA polymerase II transcription factor binding | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 2 | 33 |
| GO:0001104 | RNA polymerase II transcription cofactor activity | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 8 | 33 |
| GO:0016592 | mediator complex | 2.16e-02 | 1.00e+00 | 5.517 | 1 | 10 | 35 |
| GO:0071560 | cellular response to transforming growth factor beta stimulus | 2.22e-02 | 1.00e+00 | 5.476 | 1 | 3 | 36 |
| GO:0071407 | cellular response to organic cyclic compound | 2.35e-02 | 1.00e+00 | 5.398 | 1 | 3 | 38 |
| GO:0035019 | somatic stem cell maintenance | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 6 | 40 |
| GO:0005654 | nucleoplasm | 2.51e-02 | 1.00e+00 | 2.152 | 3 | 64 | 1082 |
| GO:0035914 | skeletal muscle cell differentiation | 2.53e-02 | 1.00e+00 | 5.289 | 1 | 2 | 41 |
| GO:0035690 | cellular response to drug | 3.02e-02 | 1.00e+00 | 5.031 | 1 | 3 | 49 |
| GO:0009611 | response to wounding | 3.20e-02 | 1.00e+00 | 4.946 | 1 | 5 | 52 |
| GO:0005515 | protein binding | 3.23e-02 | 1.00e+00 | 0.897 | 7 | 198 | 6024 |
| GO:0002039 | p53 binding | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 4 | 55 |
| GO:0043627 | response to estrogen | 3.44e-02 | 1.00e+00 | 4.839 | 1 | 5 | 56 |
| GO:0030097 | hemopoiesis | 3.44e-02 | 1.00e+00 | 4.839 | 1 | 1 | 56 |
| GO:0071356 | cellular response to tumor necrosis factor | 3.50e-02 | 1.00e+00 | 4.813 | 1 | 1 | 57 |
| GO:0010976 | positive regulation of neuron projection development | 3.56e-02 | 1.00e+00 | 4.788 | 1 | 1 | 58 |
| GO:0006310 | DNA recombination | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 7 | 65 |
| GO:0071260 | cellular response to mechanical stimulus | 3.98e-02 | 1.00e+00 | 4.624 | 1 | 4 | 65 |
| GO:0010468 | regulation of gene expression | 4.17e-02 | 1.00e+00 | 4.559 | 1 | 2 | 68 |
| GO:0003729 | mRNA binding | 4.41e-02 | 1.00e+00 | 4.476 | 1 | 4 | 72 |
| GO:0042826 | histone deacetylase binding | 4.41e-02 | 1.00e+00 | 4.476 | 1 | 4 | 72 |
| GO:0000785 | chromatin | 4.47e-02 | 1.00e+00 | 4.456 | 1 | 6 | 73 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 4.53e-02 | 1.00e+00 | 4.437 | 1 | 4 | 74 |
| GO:0002020 | protease binding | 4.53e-02 | 1.00e+00 | 4.437 | 1 | 6 | 74 |
| GO:0071222 | cellular response to lipopolysaccharide | 4.94e-02 | 1.00e+00 | 4.306 | 1 | 4 | 81 |
| GO:0009653 | anatomical structure morphogenesis | 5.06e-02 | 1.00e+00 | 4.271 | 1 | 2 | 83 |
| GO:0050821 | protein stabilization | 5.36e-02 | 1.00e+00 | 4.187 | 1 | 3 | 88 |
| GO:0071456 | cellular response to hypoxia | 5.60e-02 | 1.00e+00 | 4.123 | 1 | 3 | 92 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 6.73e-02 | 1.00e+00 | 2.197 | 2 | 39 | 699 |
| GO:0006325 | chromatin organization | 7.13e-02 | 1.00e+00 | 3.764 | 1 | 12 | 118 |
| GO:0044255 | cellular lipid metabolic process | 8.00e-02 | 1.00e+00 | 3.591 | 1 | 2 | 133 |
| GO:0010628 | positive regulation of gene expression | 8.75e-02 | 1.00e+00 | 3.456 | 1 | 7 | 146 |
| GO:0006974 | cellular response to DNA damage stimulus | 9.04e-02 | 1.00e+00 | 3.408 | 1 | 7 | 151 |
| GO:0016607 | nuclear speck | 1.02e-01 | 1.00e+00 | 3.220 | 1 | 12 | 172 |
| GO:0007049 | cell cycle | 1.04e-01 | 1.00e+00 | 3.203 | 1 | 6 | 174 |
| GO:0031625 | ubiquitin protein ligase binding | 1.06e-01 | 1.00e+00 | 3.170 | 1 | 5 | 178 |
| GO:0003714 | transcription corepressor activity | 1.06e-01 | 1.00e+00 | 3.170 | 1 | 11 | 178 |
| GO:0008380 | RNA splicing | 1.34e-01 | 1.00e+00 | 2.813 | 1 | 22 | 228 |
| GO:0044822 | poly(A) RNA binding | 1.37e-01 | 1.00e+00 | 1.602 | 2 | 50 | 1056 |
| GO:0008134 | transcription factor binding | 1.41e-01 | 1.00e+00 | 2.727 | 1 | 18 | 242 |
| GO:0043025 | neuronal cell body | 1.43e-01 | 1.00e+00 | 2.710 | 1 | 9 | 245 |
| GO:0006281 | DNA repair | 1.52e-01 | 1.00e+00 | 2.618 | 1 | 14 | 261 |
| GO:0043065 | positive regulation of apoptotic process | 1.55e-01 | 1.00e+00 | 2.580 | 1 | 7 | 268 |
| GO:0006954 | inflammatory response | 1.60e-01 | 1.00e+00 | 2.532 | 1 | 6 | 277 |
| GO:0005737 | cytoplasm | 1.87e-01 | 1.00e+00 | 0.767 | 4 | 124 | 3767 |
| GO:0043565 | sequence-specific DNA binding | 1.96e-01 | 1.00e+00 | 2.212 | 1 | 11 | 346 |
| GO:0005925 | focal adhesion | 2.07e-01 | 1.00e+00 | 2.130 | 1 | 9 | 366 |
| GO:0008284 | positive regulation of cell proliferation | 2.15e-01 | 1.00e+00 | 2.069 | 1 | 10 | 382 |
| GO:0042802 | identical protein binding | 2.65e-01 | 1.00e+00 | 1.727 | 1 | 19 | 484 |
| GO:0016032 | viral process | 2.88e-01 | 1.00e+00 | 1.585 | 1 | 26 | 534 |
| GO:0005739 | mitochondrion | 4.76e-01 | 1.00e+00 | 0.683 | 1 | 23 | 998 |
| GO:0005829 | cytosol | 4.79e-01 | 1.00e+00 | 0.361 | 2 | 86 | 2496 |
| GO:0005886 | plasma membrane | 4.98e-01 | 1.00e+00 | 0.312 | 2 | 49 | 2582 |
| GO:0044281 | small molecule metabolic process | 5.46e-01 | 1.00e+00 | 0.404 | 1 | 20 | 1211 |
| GO:0046872 | metal ion binding | 5.75e-01 | 1.00e+00 | 0.294 | 1 | 29 | 1307 |
| GO:0006351 | transcription, DNA-templated | 6.14e-01 | 1.00e+00 | 0.148 | 1 | 57 | 1446 |
| GO:0016020 | membrane | 6.72e-01 | 1.00e+00 | -0.069 | 1 | 46 | 1681 |