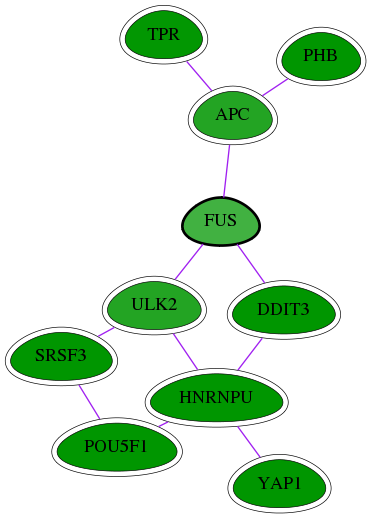
| GO:0005654 | nucleoplasm | 3.84e-08 | 5.54e-04 | 3.415 | 8 | 64 | 1082 |
| GO:0044212 | transcription regulatory region DNA binding | 3.53e-06 | 5.09e-02 | 5.102 | 4 | 17 | 168 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 3.36e-05 | 4.85e-01 | 7.794 | 2 | 2 | 13 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.28e-04 | 1.00e+00 | 4.841 | 3 | 7 | 151 |
| GO:0005515 | protein binding | 1.61e-04 | 1.00e+00 | 1.260 | 10 | 198 | 6024 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.63e-04 | 1.00e+00 | 4.722 | 3 | 15 | 164 |
| GO:0007094 | mitotic spindle assembly checkpoint | 1.99e-04 | 1.00e+00 | 6.540 | 2 | 3 | 31 |
| GO:0042594 | response to starvation | 1.99e-04 | 1.00e+00 | 6.540 | 2 | 4 | 31 |
| GO:0035019 | somatic stem cell maintenance | 3.33e-04 | 1.00e+00 | 6.172 | 2 | 6 | 40 |
| GO:0044822 | poly(A) RNA binding | 3.85e-04 | 1.00e+00 | 2.772 | 5 | 50 | 1056 |
| GO:0008380 | RNA splicing | 4.31e-04 | 1.00e+00 | 4.246 | 3 | 22 | 228 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:1901673 | regulation of spindle assembly involved in mitosis | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0043578 | nuclear matrix organization | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:2000016 | negative regulation of determination of dorsal identity | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0051171 | regulation of nitrogen compound metabolic process | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0010965 | regulation of mitotic sister chromatid separation | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0006404 | RNA import into nucleus | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0031453 | positive regulation of heterochromatin assembly | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0042483 | negative regulation of odontogenesis | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0010467 | gene expression | 7.70e-04 | 1.00e+00 | 3.108 | 4 | 45 | 669 |
| GO:0000776 | kinetochore | 8.26e-04 | 1.00e+00 | 5.517 | 2 | 5 | 63 |
| GO:0000189 | MAPK import into nucleus | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0060242 | contact inhibition | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0031990 | mRNA export from nucleus in response to heat stress | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 2 | 2 |
| GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0070840 | dynein complex binding | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0009798 | axis specification | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0046832 | negative regulation of RNA export from nucleus | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0044324 | regulation of transcription involved in anterior/posterior axis specification | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0003723 | RNA binding | 1.40e-03 | 1.00e+00 | 3.661 | 3 | 18 | 342 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1.53e-03 | 1.00e+00 | 5.068 | 2 | 8 | 86 |
| GO:0001649 | osteoblast differentiation | 1.75e-03 | 1.00e+00 | 4.971 | 2 | 5 | 92 |
| GO:0006405 | RNA export from nucleus | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 2 | 3 |
| GO:0042306 | regulation of protein import into nucleus | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 2 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0005634 | nucleus | 2.32e-03 | 1.00e+00 | 1.340 | 8 | 159 | 4559 |
| GO:0034273 | ATG1/UKL1 signaling complex | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0006999 | nuclear pore organization | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0044615 | nuclear pore nuclear basket | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0019887 | protein kinase regulator activity | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0007050 | cell cycle arrest | 3.15e-03 | 1.00e+00 | 4.540 | 2 | 5 | 124 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.40e-03 | 1.00e+00 | 2.533 | 4 | 43 | 997 |
| GO:0001714 | endodermal cell fate specification | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 2 | 5 |
| GO:0043620 | regulation of DNA-templated transcription in response to stress | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 2 | 5 |
| GO:0010793 | regulation of mRNA export from nucleus | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0048671 | negative regulation of collateral sprouting | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0070849 | response to epidermal growth factor | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0030858 | positive regulation of epithelial cell differentiation | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0050847 | progesterone receptor signaling pathway | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 2 | 6 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 3 | 6 |
| GO:0005737 | cytoplasm | 4.57e-03 | 1.00e+00 | 1.422 | 7 | 124 | 3767 |
| GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0046825 | regulation of protein export from nucleus | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0071354 | cellular response to interleukin-6 | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 2 | 7 |
| GO:0045670 | regulation of osteoclast differentiation | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 2 | 8 |
| GO:0051292 | nuclear pore complex assembly | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 4 | 8 |
| GO:0001955 | blood vessel maturation | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0035457 | cellular response to interferon-alpha | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0045667 | regulation of osteoblast differentiation | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0042405 | nuclear inclusion body | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 2 | 9 |
| GO:0006983 | ER overload response | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0043495 | protein anchor | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0003714 | transcription corepressor activity | 6.39e-03 | 1.00e+00 | 4.018 | 2 | 11 | 178 |
| GO:0035413 | positive regulation of catenin import into nucleus | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 2 | 10 |
| GO:0031274 | positive regulation of pseudopodium assembly | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 2 | 10 |
| GO:0030877 | beta-catenin destruction complex | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 2 | 10 |
| GO:0003677 | DNA binding | 7.02e-03 | 1.00e+00 | 2.244 | 4 | 49 | 1218 |
| GO:0031116 | positive regulation of microtubule polymerization | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 1 | 11 |
| GO:0051010 | microtubule plus-end binding | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 1 | 11 |
| GO:0045662 | negative regulation of myoblast differentiation | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 3 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 4 | 11 |
| GO:0045295 | gamma-catenin binding | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0032886 | regulation of microtubule-based process | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0045947 | negative regulation of translational initiation | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 12 |
| GO:0034045 | pre-autophagosomal structure membrane | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 2 | 12 |
| GO:0034399 | nuclear periphery | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 5 | 12 |
| GO:0035198 | miRNA binding | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 3 | 13 |
| GO:0044295 | axonal growth cone | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0071480 | cellular response to gamma radiation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0051019 | mitogen-activated protein kinase binding | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 13 |
| GO:0043409 | negative regulation of MAPK cascade | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 13 |
| GO:0005868 | cytoplasmic dynein complex | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0046827 | positive regulation of protein export from nucleus | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0035371 | microtubule plus-end | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 14 |
| GO:0005487 | nucleocytoplasmic transporter activity | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 3 | 14 |
| GO:0042307 | positive regulation of protein import into nucleus | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 4 | 15 |
| GO:0001824 | blastocyst development | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 15 |
| GO:0007026 | negative regulation of microtubule depolymerization | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 16 |
| GO:0046716 | muscle cell cellular homeostasis | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 16 |
| GO:0043274 | phospholipase binding | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 1 | 16 |
| GO:0051276 | chromosome organization | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 2 | 16 |
| GO:0008134 | transcription factor binding | 1.15e-02 | 1.00e+00 | 3.575 | 2 | 18 | 242 |
| GO:0090316 | positive regulation of intracellular protein transport | 1.17e-02 | 1.00e+00 | 6.407 | 1 | 2 | 17 |
| GO:0031122 | cytoplasmic microtubule organization | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 18 |
| GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1.31e-02 | 1.00e+00 | 6.246 | 1 | 2 | 19 |
| GO:0000166 | nucleotide binding | 1.31e-02 | 1.00e+00 | 3.477 | 2 | 13 | 259 |
| GO:0032757 | positive regulation of interleukin-8 production | 1.38e-02 | 1.00e+00 | 6.172 | 1 | 1 | 20 |
| GO:0009954 | proximal/distal pattern formation | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 21 |
| GO:0048675 | axon extension | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 2 | 21 |
| GO:0016575 | histone deacetylation | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 3 | 21 |
| GO:0000281 | mitotic cytokinesis | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 2 | 21 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 2 | 22 |
| GO:0010506 | regulation of autophagy | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 2 | 23 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 1 | 23 |
| GO:0045296 | cadherin binding | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 3 | 24 |
| GO:0006611 | protein export from nucleus | 1.72e-02 | 1.00e+00 | 5.850 | 1 | 3 | 25 |
| GO:0035329 | hippo signaling | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 5 | 26 |
| GO:0000045 | autophagic vacuole assembly | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 2 | 26 |
| GO:0051209 | release of sequestered calcium ion into cytosol | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 26 |
| GO:0005829 | cytosol | 1.81e-02 | 1.00e+00 | 1.531 | 5 | 86 | 2496 |
| GO:0015631 | tubulin binding | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 2 | 27 |
| GO:0034605 | cellular response to heat | 1.93e-02 | 1.00e+00 | 5.687 | 1 | 1 | 28 |
| GO:0070830 | tight junction assembly | 1.93e-02 | 1.00e+00 | 5.687 | 1 | 1 | 28 |
| GO:0071897 | DNA biosynthetic process | 1.93e-02 | 1.00e+00 | 5.687 | 1 | 1 | 28 |
| GO:0005913 | cell-cell adherens junction | 1.99e-02 | 1.00e+00 | 5.636 | 1 | 3 | 29 |
| GO:0072686 | mitotic spindle | 1.99e-02 | 1.00e+00 | 5.636 | 1 | 3 | 29 |
| GO:0006606 | protein import into nucleus | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 2 | 30 |
| GO:0031647 | regulation of protein stability | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 4 | 30 |
| GO:0010827 | regulation of glucose transport | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 4 | 30 |
| GO:0009953 | dorsal/ventral pattern formation | 2.13e-02 | 1.00e+00 | 5.540 | 1 | 2 | 31 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 2.20e-02 | 1.00e+00 | 5.494 | 1 | 2 | 32 |
| GO:0031072 | heat shock protein binding | 2.27e-02 | 1.00e+00 | 5.450 | 1 | 2 | 33 |
| GO:0033077 | T cell differentiation in thymus | 2.27e-02 | 1.00e+00 | 5.450 | 1 | 1 | 33 |
| GO:0043565 | sequence-specific DNA binding | 2.27e-02 | 1.00e+00 | 3.060 | 2 | 11 | 346 |
| GO:0008285 | negative regulation of cell proliferation | 2.36e-02 | 1.00e+00 | 3.031 | 2 | 10 | 353 |
| GO:0001942 | hair follicle development | 2.40e-02 | 1.00e+00 | 5.365 | 1 | 2 | 35 |
| GO:0007077 | mitotic nuclear envelope disassembly | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 4 | 36 |
| GO:0048538 | thymus development | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 2 | 36 |
| GO:0008645 | hexose transport | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 4 | 39 |
| GO:0051781 | positive regulation of cell division | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 1 | 39 |
| GO:0005881 | cytoplasmic microtubule | 2.74e-02 | 1.00e+00 | 5.172 | 1 | 1 | 40 |
| GO:0045785 | positive regulation of cell adhesion | 2.74e-02 | 1.00e+00 | 5.172 | 1 | 2 | 40 |
| GO:0019898 | extrinsic component of membrane | 2.88e-02 | 1.00e+00 | 5.102 | 1 | 2 | 42 |
| GO:0031124 | mRNA 3'-end processing | 2.88e-02 | 1.00e+00 | 5.102 | 1 | 2 | 42 |
| GO:0006369 | termination of RNA polymerase II transcription | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 2 | 45 |
| GO:0043525 | positive regulation of neuron apoptotic process | 3.14e-02 | 1.00e+00 | 4.971 | 1 | 4 | 46 |
| GO:0016328 | lateral plasma membrane | 3.14e-02 | 1.00e+00 | 4.971 | 1 | 3 | 46 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.16e-02 | 1.00e+00 | 2.804 | 2 | 24 | 413 |
| GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 3.21e-02 | 1.00e+00 | 4.940 | 1 | 2 | 47 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.25e-02 | 1.00e+00 | 2.783 | 2 | 30 | 419 |
| GO:0006986 | response to unfolded protein | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 4 | 50 |
| GO:0045732 | positive regulation of protein catabolic process | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 1 | 50 |
| GO:0034976 | response to endoplasmic reticulum stress | 3.48e-02 | 1.00e+00 | 4.822 | 1 | 2 | 51 |
| GO:0045454 | cell redox homeostasis | 3.48e-02 | 1.00e+00 | 4.822 | 1 | 1 | 51 |
| GO:0060041 | retina development in camera-type eye | 3.48e-02 | 1.00e+00 | 4.822 | 1 | 2 | 51 |
| GO:0009611 | response to wounding | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 5 | 52 |
| GO:0015758 | glucose transport | 3.75e-02 | 1.00e+00 | 4.713 | 1 | 4 | 55 |
| GO:0005643 | nuclear pore | 3.88e-02 | 1.00e+00 | 4.661 | 1 | 11 | 57 |
| GO:0006396 | RNA processing | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 5 | 59 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 6 | 59 |
| GO:0008013 | beta-catenin binding | 4.08e-02 | 1.00e+00 | 4.587 | 1 | 9 | 60 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 4.14e-02 | 1.00e+00 | 2.593 | 2 | 24 | 478 |
| GO:0006406 | mRNA export from nucleus | 4.15e-02 | 1.00e+00 | 4.563 | 1 | 4 | 61 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 4.15e-02 | 1.00e+00 | 4.563 | 1 | 2 | 61 |
| GO:0007409 | axonogenesis | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 2 | 65 |
| GO:0010468 | regulation of gene expression | 4.62e-02 | 1.00e+00 | 4.407 | 1 | 2 | 68 |
| GO:0032587 | ruffle membrane | 4.75e-02 | 1.00e+00 | 4.365 | 1 | 3 | 70 |
| GO:0042826 | histone deacetylase binding | 4.88e-02 | 1.00e+00 | 4.324 | 1 | 4 | 72 |
| GO:0003729 | mRNA binding | 4.88e-02 | 1.00e+00 | 4.324 | 1 | 4 | 72 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 4 | 74 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 1 | 75 |
| GO:0060070 | canonical Wnt signaling pathway | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 4 | 75 |
| GO:0071013 | catalytic step 2 spliceosome | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 7 | 78 |
| GO:0001822 | kidney development | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 3 | 78 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 5.35e-02 | 1.00e+00 | 4.190 | 1 | 2 | 79 |
| GO:0009653 | anatomical structure morphogenesis | 5.61e-02 | 1.00e+00 | 4.119 | 1 | 2 | 83 |
| GO:0009952 | anterior/posterior pattern specification | 5.61e-02 | 1.00e+00 | 4.119 | 1 | 4 | 83 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 5.69e-02 | 1.00e+00 | 2.339 | 2 | 37 | 570 |
| GO:0005923 | tight junction | 5.81e-02 | 1.00e+00 | 4.068 | 1 | 2 | 86 |
| GO:0005770 | late endosome | 6.27e-02 | 1.00e+00 | 3.955 | 1 | 2 | 93 |
| GO:0030659 | cytoplasmic vesicle membrane | 6.79e-02 | 1.00e+00 | 3.836 | 1 | 2 | 101 |
| GO:0030308 | negative regulation of cell growth | 7.37e-02 | 1.00e+00 | 3.713 | 1 | 6 | 110 |
| GO:0006461 | protein complex assembly | 7.44e-02 | 1.00e+00 | 3.700 | 1 | 4 | 111 |
| GO:0030529 | ribonucleoprotein complex | 7.50e-02 | 1.00e+00 | 3.687 | 1 | 5 | 112 |
| GO:0005635 | nuclear envelope | 7.57e-02 | 1.00e+00 | 3.674 | 1 | 5 | 113 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 8.15e-02 | 1.00e+00 | 2.045 | 2 | 39 | 699 |
| GO:0030027 | lamellipodium | 8.34e-02 | 1.00e+00 | 3.528 | 1 | 7 | 125 |
| GO:0030335 | positive regulation of cell migration | 8.47e-02 | 1.00e+00 | 3.505 | 1 | 5 | 127 |
| GO:0016477 | cell migration | 8.53e-02 | 1.00e+00 | 3.494 | 1 | 4 | 128 |
| GO:0005215 | transporter activity | 8.66e-02 | 1.00e+00 | 3.472 | 1 | 3 | 130 |
| GO:0016055 | Wnt signaling pathway | 9.17e-02 | 1.00e+00 | 3.386 | 1 | 4 | 138 |
| GO:0008017 | microtubule binding | 9.74e-02 | 1.00e+00 | 3.294 | 1 | 2 | 147 |
| GO:0042981 | regulation of apoptotic process | 9.93e-02 | 1.00e+00 | 3.265 | 1 | 4 | 150 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 1.00e-01 | 1.00e+00 | 1.870 | 2 | 41 | 789 |
| GO:0046777 | protein autophosphorylation | 1.04e-01 | 1.00e+00 | 3.190 | 1 | 4 | 158 |
| GO:0031965 | nuclear membrane | 1.11e-01 | 1.00e+00 | 3.102 | 1 | 10 | 168 |
| GO:0016607 | nuclear speck | 1.13e-01 | 1.00e+00 | 3.068 | 1 | 12 | 172 |
| GO:0005667 | transcription factor complex | 1.15e-01 | 1.00e+00 | 3.043 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.17e-01 | 1.00e+00 | 3.018 | 1 | 5 | 178 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.21e-01 | 1.00e+00 | 2.971 | 1 | 22 | 184 |
| GO:0007165 | signal transduction | 1.27e-01 | 1.00e+00 | 1.669 | 2 | 13 | 907 |
| GO:0019221 | cytokine-mediated signaling pathway | 1.43e-01 | 1.00e+00 | 2.706 | 1 | 8 | 221 |
| GO:0007067 | mitotic nuclear division | 1.47e-01 | 1.00e+00 | 2.668 | 1 | 6 | 227 |
| GO:0003713 | transcription coactivator activity | 1.53e-01 | 1.00e+00 | 2.605 | 1 | 24 | 237 |
| GO:0005975 | carbohydrate metabolic process | 1.62e-01 | 1.00e+00 | 2.511 | 1 | 8 | 253 |
| GO:0043065 | positive regulation of apoptotic process | 1.71e-01 | 1.00e+00 | 2.428 | 1 | 7 | 268 |
| GO:0005743 | mitochondrial inner membrane | 1.76e-01 | 1.00e+00 | 2.380 | 1 | 6 | 277 |
| GO:0019899 | enzyme binding | 1.76e-01 | 1.00e+00 | 2.380 | 1 | 9 | 277 |
| GO:0004674 | protein serine/threonine kinase activity | 1.94e-01 | 1.00e+00 | 2.227 | 1 | 6 | 308 |
| GO:0019901 | protein kinase binding | 1.99e-01 | 1.00e+00 | 2.186 | 1 | 18 | 317 |
| GO:0005813 | centrosome | 2.04e-01 | 1.00e+00 | 2.145 | 1 | 7 | 326 |
| GO:0003682 | chromatin binding | 2.05e-01 | 1.00e+00 | 2.141 | 1 | 19 | 327 |
| GO:0008283 | cell proliferation | 2.06e-01 | 1.00e+00 | 2.137 | 1 | 14 | 328 |
| GO:0005524 | ATP binding | 2.25e-01 | 1.00e+00 | 1.152 | 2 | 31 | 1298 |
| GO:0007155 | cell adhesion | 2.28e-01 | 1.00e+00 | 1.967 | 1 | 5 | 369 |
| GO:0046982 | protein heterodimerization activity | 2.34e-01 | 1.00e+00 | 1.924 | 1 | 8 | 380 |
| GO:0000278 | mitotic cell cycle | 2.40e-01 | 1.00e+00 | 1.883 | 1 | 15 | 391 |
| GO:0009986 | cell surface | 2.46e-01 | 1.00e+00 | 1.843 | 1 | 9 | 402 |
| GO:0055085 | transmembrane transport | 2.64e-01 | 1.00e+00 | 1.729 | 1 | 8 | 435 |
| GO:0044267 | cellular protein metabolic process | 2.84e-01 | 1.00e+00 | 1.605 | 1 | 20 | 474 |
| GO:0042802 | identical protein binding | 2.89e-01 | 1.00e+00 | 1.575 | 1 | 19 | 484 |
| GO:0016032 | viral process | 3.14e-01 | 1.00e+00 | 1.433 | 1 | 26 | 534 |
| GO:0006915 | apoptotic process | 3.25e-01 | 1.00e+00 | 1.378 | 1 | 12 | 555 |
| GO:0016020 | membrane | 3.28e-01 | 1.00e+00 | 0.779 | 2 | 46 | 1681 |
| GO:0042803 | protein homodimerization activity | 3.44e-01 | 1.00e+00 | 1.277 | 1 | 19 | 595 |
| GO:0005887 | integral component of plasma membrane | 4.70e-01 | 1.00e+00 | 0.700 | 1 | 10 | 888 |
| GO:0005739 | mitochondrion | 5.12e-01 | 1.00e+00 | 0.531 | 1 | 23 | 998 |
| GO:0008270 | zinc ion binding | 5.12e-01 | 1.00e+00 | 0.533 | 1 | 27 | 997 |
| GO:0044281 | small molecule metabolic process | 5.84e-01 | 1.00e+00 | 0.252 | 1 | 20 | 1211 |
| GO:0005730 | nucleolus | 7.01e-01 | 1.00e+00 | -0.186 | 1 | 66 | 1641 |
| GO:0070062 | extracellular vesicular exosome | 8.38e-01 | 1.00e+00 | -0.735 | 1 | 57 | 2400 |
| GO:0005886 | plasma membrane | 8.61e-01 | 1.00e+00 | -0.840 | 1 | 49 | 2582 |