meta-reg-snw-4869
Interacting subnets
| Subnetwork | Dataset | Score | p-value 1 | p-value 2 | p-value 3 | Size | Highlighted genes |
|---|---|---|---|---|---|---|---|
| int-snw-2023 | wolf-screen-ratio-mammosphere-adherent | 0.930 | 2.50e-15 | 2.74e-03 | 4.31e-02 | 28 | 26 |
| reg-snw-4869 | wolf-screen-ratio-mammosphere-adherent | 0.828 | 9.35e-07 | 2.92e-03 | 5.55e-03 | 13 | 12 |
| int-snw-2957 | wolf-screen-ratio-mammosphere-adherent | 0.941 | 9.03e-16 | 2.14e-03 | 3.61e-02 | 25 | 21 |
| int-snw-811 | wolf-screen-ratio-mammosphere-adherent | 0.932 | 2.10e-15 | 2.63e-03 | 4.19e-02 | 24 | 22 |
| int-snw-3305 | wolf-screen-ratio-mammosphere-adherent | 0.942 | 8.57e-16 | 2.11e-03 | 3.57e-02 | 23 | 22 |
| int-snw-57761 | wolf-screen-ratio-mammosphere-adherent | 0.969 | 7.13e-17 | 1.14e-03 | 2.28e-02 | 16 | 14 |
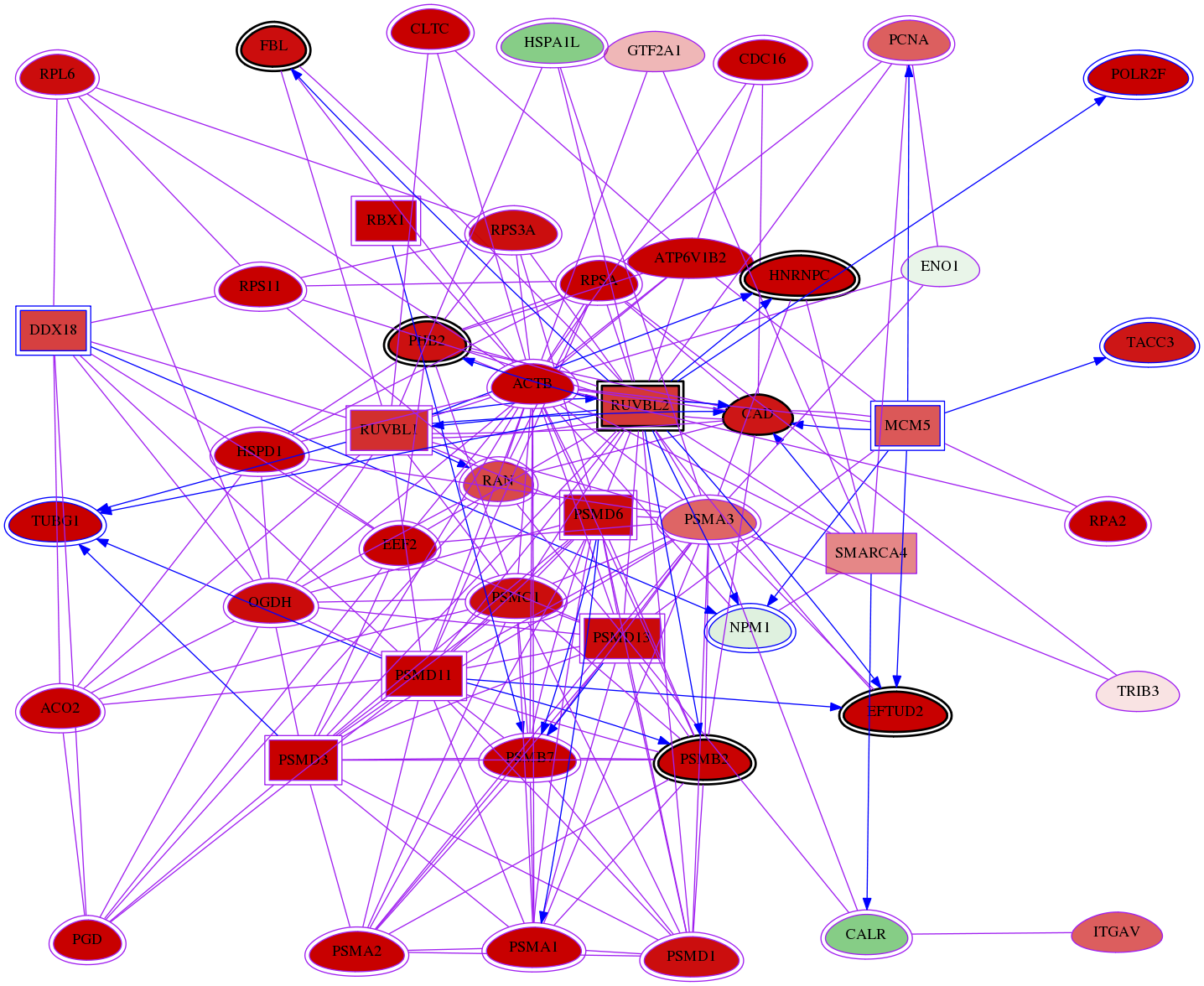
Genes (48)
| Gene Symbol | Entrez Gene ID | Frequency | wolf-screen-ratio-mammosphere-adherent gene score | Best subnetwork score | Degree | wolf adherent-list Hits GI | wolf mammosphere no adherent-list Hits GI |
|---|---|---|---|---|---|---|---|
| PSMA3 | 5684 | 90 | 0.533 | 0.815 | 238 | Yes | - |
| DDX18 | 8886 | 25 | 0.660 | 1.013 | 215 | Yes | - |
| PSMA2 | 5683 | 112 | 1.093 | 1.106 | 108 | Yes | - |
| PHB2 | 11331 | 43 | 0.829 | 0.956 | 151 | Yes | - |
| POLR2F | 5435 | 62 | 0.891 | 0.956 | 26 | Yes | - |
| SMARCA4 | 6597 | 26 | 0.416 | 0.941 | 253 | - | - |
| NPM1 | 4869 | 6 | -0.112 | 0.828 | 62 | Yes | - |
| ATP6V1B2 | 526 | 53 | 0.881 | 1.076 | 278 | - | - |
| RPA2 | 6118 | 96 | 1.250 | 1.151 | 76 | Yes | - |
| CAD | 790 | 91 | 0.807 | 0.973 | 400 | - | - |
| CALR | 811 | 38 | -0.418 | 0.932 | 79 | - | Yes |
| PCNA | 5111 | 33 | 0.553 | 0.974 | 294 | Yes | - |
| PSMD1 | 5707 | 86 | 0.836 | 0.830 | 118 | Yes | - |
| EEF2 | 1938 | 64 | 0.890 | 1.043 | 301 | Yes | - |
| RPS11 | 6205 | 62 | 0.993 | 1.113 | 175 | Yes | - |
| MCM5 | 4174 | 23 | 0.578 | 0.830 | 273 | Yes | - |
| EFTUD2 | 9343 | 93 | 0.883 | 0.956 | 108 | Yes | - |
| TUBG1 | 7283 | 98 | 0.974 | 0.973 | 91 | Yes | - |
| PSMD11 | 5717 | 124 | 1.095 | 1.106 | 218 | Yes | - |
| RAN | 5901 | 89 | 0.632 | 0.899 | 258 | Yes | - |
| FBL | 2091 | 42 | 0.839 | 0.956 | 79 | Yes | - |
| HSPD1 | 3329 | 70 | 0.913 | 1.035 | 325 | Yes | - |
| ENO1 | 2023 | 28 | -0.078 | 0.930 | 180 | - | - |
| TACC3 | 10460 | 8 | 0.806 | 0.830 | 35 | Yes | - |
| RBX1 | 9978 | 115 | 1.185 | 0.934 | 148 | Yes | - |
| PSMB7 | 5695 | 118 | 0.982 | 0.934 | 90 | Yes | - |
| HNRNPC | 3183 | 108 | 1.812 | 0.973 | 181 | Yes | - |
| RPL6 | 6128 | 37 | 0.844 | 1.113 | 164 | Yes | - |
| PSMA1 | 5682 | 100 | 0.996 | 0.878 | 152 | Yes | - |
| OGDH | 4967 | 72 | 0.847 | 0.802 | 126 | Yes | - |
| RPSA | 3921 | 120 | 1.327 | 1.151 | 152 | Yes | - |
| RPS3A | 6189 | 40 | 0.835 | 1.069 | 166 | Yes | - |
| PGD | 5226 | 89 | 1.201 | 1.106 | 152 | Yes | - |
| HSPA1L | 3305 | 34 | -0.414 | 0.942 | 125 | - | Yes |
| TRIB3 | 57761 | 26 | 0.097 | 0.969 | 31 | - | - |
| ACO2 | 50 | 65 | 1.000 | 1.076 | 191 | Yes | - |
| CDC16 | 8881 | 53 | 0.950 | 1.020 | 80 | Yes | - |
| RUVBL1 | 8607 | 95 | 0.720 | 0.973 | 469 | Yes | - |
| PSMD13 | 5719 | 56 | 0.848 | 0.801 | 114 | Yes | - |
| GTF2A1 | 2957 | 26 | 0.251 | 0.941 | 52 | - | - |
| PSMD3 | 5709 | 100 | 0.986 | 1.106 | 201 | Yes | - |
| CLTC | 1213 | 56 | 0.884 | 1.138 | 247 | Yes | - |
| PSMB2 | 5690 | 116 | 0.877 | 0.956 | 169 | Yes | - |
| ACTB | 60 | 134 | 1.153 | 1.151 | 610 | Yes | - |
| ITGAV | 3685 | 39 | 0.556 | 0.949 | 37 | - | - |
| RUVBL2 | 10856 | 95 | 0.693 | 0.956 | 532 | Yes | - |
| PSMD6 | 9861 | 79 | 0.848 | 0.878 | 143 | Yes | - |
| PSMC1 | 5700 | 51 | 0.840 | 1.018 | 137 | Yes | - |
Interactions (208)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| PSMA3 | 5684 | PSMB2 | 5690 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Mouse, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, Krogan_Core |
| PSMD11 | 5717 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastHigh, BioGrid, INTEROLOG |
| RPS11 | 6205 | RUVBL2 | 10856 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: IntAct, BioGrid |
| HNRNPC | 3183 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid |
| CAD | 790 | MCM5 | 4174 | pd | < | reg.ITFP.txt: no annot |
| CAD | 790 | MCM5 | 4174 | pp | -- | int.I2D: IntAct_Yeast |
| EEF2 | 1938 | PGD | 5226 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA1 | 5682 | PSMD13 | 5719 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: IntAct, YeastMedium, BioGrid, BioGrid_Yeast, IntAct_Yeast |
| PSMA1 | 5682 | PSMB2 | 5690 | pp | -- | int.Intact: MI:0915(physical association), MI:0914(association); int.I2D: HPRD, IntAct, IntAct_Mouse, IntAct_Yeast, INTEROLOG, BioGrid_Yeast, Krogan_Core, MINT_Yeast, YeastHigh; int.HPRD: yeast 2-hybrid |
| EEF2 | 1938 | HSPD1 | 3329 | pp | -- | int.I2D: YeastLow, YeastMedium |
| PSMD11 | 5717 | RUVBL1 | 8607 | pp | -- | int.I2D: YeastLow |
| PSMA2 | 5683 | PSMD13 | 5719 | pp | -- | int.I2D: YeastLow |
| ACO2 | 50 | OGDH | 4967 | pp | -- | int.I2D: YeastLow |
| CAD | 790 | CDC16 | 8881 | pp | -- | int.I2D: IntAct_Yeast |
| EFTUD2 | 9343 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| EFTUD2 | 9343 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid |
| ACTB | 60 | GTF2A1 | 2957 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMD1 | 5707 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, MINT_Yeast, Krogan_Core |
| ACTB | 60 | CLTC | 1213 | pp | -- | int.I2D: IntAct_Yeast |
| ACTB | 60 | RUVBL1 | 8607 | pp | -- | int.I2D: IntAct_Yeast, BioGrid, BioGrid_Yeast |
| ACTB | 60 | HSPD1 | 3329 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, YeastLow |
| PSMB7 | 5695 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA2 | 5683 | PSMB7 | 5695 | pp | -- | int.Intact: MI:0914(association); int.I2D: BCI, BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct, Krogan_Core, MINT_Yeast, YeastLow, IntAct_Yeast |
| PSMD1 | 5707 | PSMD13 | 5719 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT_Yeast, BioGrid, BioGrid_Yeast, IntAct_Yeast, Krogan_Core |
| PSMC1 | 5700 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, MINT_Yeast, YeastHigh, Krogan_Core |
| PGD | 5226 | RAN | 5901 | pp | -- | int.I2D: YeastLow |
| ATP6V1B2 | 526 | OGDH | 4967 | pp | -- | int.I2D: IntAct_Yeast |
| RAN | 5901 | RUVBL1 | 8607 | pd | < | reg.ITFP.txt: no annot |
| RPL6 | 6128 | RPS3A | 6189 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | PSMD13 | 5719 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMB7 | 5695 | PSMC1 | 5700 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA2 | 5683 | PSMD1 | 5707 | pp | -- | int.I2D: BioGrid_Yeast |
| CAD | 790 | RUVBL1 | 8607 | pd | < | reg.ITFP.txt: no annot |
| CAD | 790 | RUVBL1 | 8607 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| PSMA3 | 5684 | PSMD13 | 5719 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: IntAct, BioGrid |
| PSMA2 | 5683 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| MCM5 | 4174 | EFTUD2 | 9343 | pd | > | reg.ITFP.txt: no annot |
| RPSA | 3921 | PSMD11 | 5717 | pp | -- | int.I2D: IntAct_Yeast |
| ACO2 | 50 | HSPD1 | 3329 | pp | -- | int.I2D: IntAct_Yeast |
| OGDH | 4967 | RUVBL1 | 8607 | pp | -- | int.I2D: YeastLow |
| RPL6 | 6128 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | PSMD6 | 9861 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| ACO2 | 50 | PSMD11 | 5717 | pp | -- | int.I2D: YeastLow |
| PSMB2 | 5690 | PSMD13 | 5719 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium |
| CALR | 811 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid |
| ACTB | 60 | PSMB2 | 5690 | pp | -- | int.I2D: BioGrid_Yeast |
| GTF2A1 | 2957 | SMARCA4 | 6597 | pp | -- | int.I2D: BioGrid_Yeast |
| HSPD1 | 3329 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| ACTB | 60 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMC1 | 5700 | PSMD13 | 5719 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, MINT_Yeast, YeastHigh, BioGrid, IntAct_Yeast, Krogan_Core |
| PSMA1 | 5682 | PSMD6 | 9861 | pd | < | reg.ITFP.txt: no annot |
| PSMA1 | 5682 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, YeastMedium |
| CALR | 811 | RAN | 5901 | pp | -- | int.I2D: IntAct_Worm, BioGrid_Worm, BIND_Worm, CORE_2, MINT_Worm |
| PSMD3 | 5709 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastHigh, INTEROLOG |
| CLTC | 1213 | RUVBL1 | 8607 | pp | -- | int.I2D: MINT_Worm, IntAct_Worm |
| ACTB | 60 | PSMC1 | 5700 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMD1 | 5707 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast |
| ATP6V1B2 | 526 | CLTC | 1213 | pp | -- | int.I2D: IntAct_Yeast |
| ACTB | 60 | EEF2 | 1938 | pp | -- | int.I2D: YeastLow, IntAct_Yeast |
| ACTB | 60 | RBX1 | 9978 | pp | -- | int.I2D: BioGrid_Yeast |
| HSPA1L | 3305 | RUVBL1 | 8607 | pp | -- | int.I2D: BioGrid |
| RPS3A | 6189 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| RAN | 5901 | RPS11 | 6205 | pp | -- | int.I2D: IntAct_Worm, BioGrid_Worm, BIND_Worm, MINT_Worm, NON_CORE |
| PSMA1 | 5682 | PSMA2 | 5683 | pp | -- | int.Intact: MI:0915(physical association), MI:0914(association); int.I2D: BioGrid_Worm, BioGrid_Yeast, BIND_Worm, CE_DATA, HPRD, IntAct, IntAct_Mouse, IntAct_Yeast, INTEROLOG, MINT_Worm, BioGrid, IntAct_Fly, IntAct_Worm, Krogan_Core, MINT_Yeast, YeastHigh; int.HPRD: yeast 2-hybrid |
| RPSA | 3921 | RAN | 5901 | pp | -- | int.I2D: IntAct_Worm, BioGrid_Worm, BIND_Worm, CORE_2, MINT_Worm |
| ACTB | 60 | RPSA | 3921 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct_Yeast, IntAct |
| PSMD6 | 9861 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow |
| ACTB | 60 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast |
| RPS3A | 6189 | RUVBL2 | 10856 | pp | -- | int.I2D: IntAct_Yeast |
| OGDH | 4967 | PSMD13 | 5719 | pp | -- | int.I2D: YeastLow |
| OGDH | 4967 | PSMD6 | 9861 | pp | -- | int.I2D: YeastLow |
| RUVBL1 | 8607 | RUVBL2 | 10856 | pd | <> | reg.ITFP.txt: no annot |
| RUVBL1 | 8607 | RUVBL2 | 10856 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association), MI:0914(association); int.I2D: BioGrid, BioGrid_Mouse, BioGrid_Yeast, BIND_Yeast, HPRD, IntAct, IntAct_Fly, IntAct_Worm, IntAct_Yeast, INTEROLOG, MINT_Worm, MINT_Yeast, BCI, BioGrid_Fly, BIND_Fly, BIND_Worm, FlyHigh, INNATEDB, Krogan_Core, MIPS, YeastHigh, Yu_GoldStd; int.Ravasi: -; int.HPRD: in vitro, in vivo, yeast 2-hybrid; int.DIP: MI:0915(physical association) |
| RPSA | 3921 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| EEF2 | 1938 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid |
| PSMA1 | 5682 | PSMB7 | 5695 | pp | -- | int.Intact: MI:0914(association); int.I2D: BioGrid_Yeast, IntAct, IntAct_Mouse, IntAct_Yeast, YeastLow, Krogan_Core, MINT_Yeast |
| ACTB | 60 | PSMA1 | 5682 | pp | -- | int.I2D: BioGrid_Yeast |
| EEF2 | 1938 | PSMD3 | 5709 | pp | -- | int.I2D: Krogan_NonCore |
| ACTB | 60 | CAD | 790 | pp | -- | int.I2D: IntAct_Yeast |
| OGDH | 4967 | PGD | 5226 | pp | -- | int.I2D: YeastLow |
| PSMD11 | 5717 | DDX18 | 8886 | pp | -- | int.I2D: YeastLow |
| PSMB7 | 5695 | RBX1 | 9978 | pd | < | reg.ITFP.txt: no annot |
| PSMA3 | 5684 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow |
| CAD | 790 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| CAD | 790 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| PGD | 5226 | PSMD11 | 5717 | pp | -- | int.I2D: YeastLow |
| HNRNPC | 3183 | RUVBL1 | 8607 | pd | < | reg.ITFP.txt: no annot |
| PSMD3 | 5709 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow |
| PSMD3 | 5709 | TUBG1 | 7283 | pd | > | reg.ITFP.txt: no annot |
| MCM5 | 4174 | PCNA | 5111 | pd | > | reg.ITFP.txt: no annot |
| NPM1 | 4869 | SMARCA4 | 6597 | pp | -- | int.I2D: BioGrid |
| RUVBL1 | 8607 | DDX18 | 8886 | pp | -- | int.I2D: YeastLow |
| PSMB7 | 5695 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast |
| ENO1 | 2023 | PCNA | 5111 | pp | -- | int.I2D: MINT, BioGrid; int.Mint: MI:0407(direct interaction), MI:0915(physical association) |
| ACTB | 60 | ENO1 | 2023 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| HNRNPC | 3183 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| ATP6V1B2 | 526 | CDC16 | 8881 | pp | -- | int.I2D: IntAct_Yeast |
| ACTB | 60 | FBL | 2091 | pp | -- | int.I2D: MINT |
| MCM5 | 4174 | RPA2 | 6118 | pp | -- | int.I2D: BioGrid, HPRD; int.HPRD: yeast 2-hybrid |
| PSMA3 | 5684 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, YeastLow |
| PSMC1 | 5700 | PSMD1 | 5707 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, Krogan_Core, MINT_Yeast |
| PSMD1 | 5707 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid, BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, IntAct, Krogan_Core |
| HNRNPC | 3183 | SMARCA4 | 6597 | pp | -- | int.I2D: BioGrid |
| PSMA3 | 5684 | PSMD1 | 5707 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast |
| EEF2 | 1938 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| CALR | 811 | SMARCA4 | 6597 | pd | < | reg.ITFP.txt: no annot |
| ACTB | 60 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA3 | 5684 | EFTUD2 | 9343 | pp | -- | int.I2D: BioGrid |
| PSMB2 | 5690 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| OGDH | 4967 | DDX18 | 8886 | pp | -- | int.I2D: YeastLow |
| HSPD1 | 3329 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid |
| MCM5 | 4174 | TACC3 | 10460 | pd | > | reg.ITFP.txt: no annot |
| PGD | 5226 | PSMD3 | 5709 | pp | -- | int.I2D: YeastLow |
| ACO2 | 50 | PSMC1 | 5700 | pp | -- | int.I2D: YeastLow |
| PSMC1 | 5700 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow |
| PSMD13 | 5719 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastHigh |
| PSMA3 | 5684 | PSMB7 | 5695 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Mouse, MINT_Yeast, YeastLow, Krogan_Core |
| PSMB2 | 5690 | PSMB7 | 5695 | pp | -- | int.Intact: MI:0914(association); int.I2D: BioGrid_Yeast, Krogan_Core, MINT_Yeast, YeastLow, IntAct |
| PSMD3 | 5709 | PSMD6 | 9861 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid_Worm, BioGrid_Yeast, BIND_Worm, CE_DATA, IntAct, IntAct_Yeast, MINT_Worm, MINT_Yeast, YeastHigh, BioGrid, BIND_Yeast, IntAct_Worm, INTEROLOG, Krogan_Core, MIPS |
| RPSA | 3921 | TRIB3 | 57761 | pp | -- | int.I2D: BioGrid |
| PCNA | 5111 | SMARCA4 | 6597 | pp | -- | int.I2D: BioGrid |
| FBL | 2091 | RUVBL1 | 8607 | pp | -- | int.I2D: BioGrid |
| MCM5 | 4174 | NPM1 | 4869 | pd | > | reg.ITFP.txt: no annot |
| RPS3A | 6189 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| TUBG1 | 7283 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| NPM1 | 4869 | DDX18 | 8886 | pd | < | reg.ITFP.txt: no annot |
| PSMA3 | 5684 | RUVBL1 | 8607 | pp | -- | int.I2D: BioGrid |
| ACTB | 60 | RPL6 | 6128 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | RAN | 5901 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, NON_CORE |
| TUBG1 | 7283 | RUVBL1 | 8607 | pd | < | reg.ITFP.txt: no annot |
| POLR2F | 5435 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| CAD | 790 | RAN | 5901 | pp | -- | int.I2D: IntAct_Yeast |
| ACTB | 60 | PSMD6 | 9861 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMC1 | 5700 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, MINT_Yeast, BioGrid, IntAct_Yeast, Krogan_Core, YeastHigh |
| PSMB2 | 5690 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium |
| PSMB7 | 5695 | PSMD1 | 5707 | pp | -- | int.Intact: MI:0914(association); int.I2D: BioGrid_Yeast, IntAct |
| PSMA1 | 5682 | PSMA3 | 5684 | pp | -- | int.Intact: MI:0914(association), MI:0915(physical association); int.I2D: BioGrid_Worm, BioGrid_Yeast, CE_DATA, HPRD, IntAct, IntAct_Fly, IntAct_Mouse, IntAct_Worm, IntAct_Yeast, MINT_Worm, BioGrid, BioGrid_Fly, BIND_Fly, BIND_Yeast, FlyHigh, Krogan_Core, MINT_Fly, MINT_Yeast, YeastHigh, Yu_GoldStd; int.HPRD: in vitro |
| RPL6 | 6128 | DDX18 | 8886 | pp | -- | int.I2D: BioGrid_Yeast |
| ATP6V1B2 | 526 | RUVBL2 | 10856 | pp | -- | int.I2D: IntAct_Yeast |
| PSMA2 | 5683 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| PSMD11 | 5717 | TUBG1 | 7283 | pd | > | reg.ITFP.txt: no annot |
| PSMC1 | 5700 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastHigh, INTEROLOG |
| RPSA | 3921 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| PSMA1 | 5682 | PSMD1 | 5707 | pp | -- | int.I2D: BioGrid_Yeast |
| EEF2 | 1938 | DDX18 | 8886 | pp | -- | int.I2D: YeastLow |
| PSMA3 | 5684 | PSMC1 | 5700 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | PSMB7 | 5695 | pp | -- | int.I2D: BioGrid_Yeast |
| MCM5 | 4174 | PSMD13 | 5719 | pp | -- | int.I2D: YeastLow |
| PSMA3 | 5684 | TRIB3 | 57761 | pp | -- | int.I2D: BioGrid |
| ACTB | 60 | RPA2 | 6118 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| OGDH | 4967 | PSMD3 | 5709 | pp | -- | int.I2D: YeastLow |
| PSMA2 | 5683 | PSMC1 | 5700 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Fly, YeastLow |
| CAD | 790 | PSMB2 | 5690 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMB7 | 5695 | PSMD6 | 9861 | pd | < | reg.ITFP.txt: no annot |
| PSMB7 | 5695 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct |
| PSMD11 | 5717 | EFTUD2 | 9343 | pd | > | reg.ITFP.txt: no annot |
| RUVBL1 | 8607 | PSMD6 | 9861 | pp | -- | int.I2D: YeastLow |
| ACO2 | 50 | PGD | 5226 | pp | -- | int.I2D: YeastLow |
| ACTB | 60 | NPM1 | 4869 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| ATP6V1B2 | 526 | RPSA | 3921 | pp | -- | int.I2D: IntAct_Yeast |
| ACTB | 60 | CDC16 | 8881 | pp | -- | int.I2D: YeastLow |
| PSMD11 | 5717 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow |
| OGDH | 4967 | PSMD11 | 5717 | pp | -- | int.I2D: YeastLow |
| CALR | 811 | ITGAV | 3685 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vitro, in vivo |
| FBL | 2091 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| FBL | 2091 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid |
| NPM1 | 4869 | RUVBL2 | 10856 | pd | < | reg.ITFP.txt: no annot |
| RPSA | 3921 | PCNA | 5111 | pp | -- | int.I2D: MINT, BioGrid; int.Mint: MI:0915(physical association) |
| ACTB | 60 | RPS3A | 6189 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| PSMB7 | 5695 | PSMD13 | 5719 | pd | < | reg.ITFP.txt: no annot |
| CAD | 790 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| PCNA | 5111 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid |
| EEF2 | 1938 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow, BioGrid_Yeast, IntAct_Yeast |
| PSMD1 | 5707 | RUVBL2 | 10856 | pp | -- | int.I2D: IntAct_Yeast |
| ACO2 | 50 | RUVBL1 | 8607 | pp | -- | int.I2D: YeastLow |
| PSMA3 | 5684 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| HSPA1L | 3305 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid |
| PSMB2 | 5690 | PSMD6 | 9861 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium |
| HSPD1 | 3329 | RPSA | 3921 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| PSMA1 | 5682 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| HSPA1L | 3305 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid |
| OGDH | 4967 | RPL6 | 6128 | pp | -- | int.I2D: IntAct_Yeast |
| PSMA3 | 5684 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast |
| OGDH | 4967 | RAN | 5901 | pp | -- | int.I2D: YeastLow |
| HSPD1 | 3329 | OGDH | 4967 | pp | -- | int.I2D: YeastLow |
| EEF2 | 1938 | PSMD13 | 5719 | pp | -- | int.I2D: YeastLow |
| PSMB2 | 5690 | PSMD11 | 5717 | pd | < | reg.ITFP.txt: no annot |
| PSMB2 | 5690 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| OGDH | 4967 | PSMC1 | 5700 | pp | -- | int.I2D: YeastLow |
| PSMA2 | 5683 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, YeastLow, BioGrid |
| PSMA1 | 5682 | PSMC1 | 5700 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct_Fly, YeastMedium, IntAct |
| PSMD11 | 5717 | PSMD13 | 5719 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, IntAct_Yeast, Krogan_Core, MINT_Yeast, Tarassov_PCA, YeastHigh, BioGrid, INTEROLOG |
| PSMD3 | 5709 | PSMD13 | 5719 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT_Yeast, YeastHigh, BioGrid, BioGrid_Yeast, IntAct_Yeast, Krogan_Core |
| SMARCA4 | 6597 | RUVBL2 | 10856 | pp | -- | int.I2D: YeastLow |
| ACTB | 60 | ATP6V1B2 | 526 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | SMARCA4 | 6597 | pp | -- | int.Intact: MI:0914(association); int.I2D: BioGrid, IntAct, BCI, HPRD; int.HPRD: in vitro, in vivo |
| PSMD13 | 5719 | PSMD6 | 9861 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid, BioGrid_Yeast, IntAct, IntAct_Yeast, MINT_Yeast, YeastHigh, Krogan_Core |
| PSMC1 | 5700 | RUVBL1 | 8607 | pp | -- | int.I2D: YeastLow |
| RPSA | 3921 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| ATP6V1B2 | 526 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| PGD | 5226 | DDX18 | 8886 | pp | -- | int.I2D: YeastLow |
| ACTB | 60 | MCM5 | 4174 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| ATP6V1B2 | 526 | MCM5 | 4174 | pp | -- | int.I2D: IntAct_Yeast |
| RPS11 | 6205 | DDX18 | 8886 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | RUVBL2 | 10856 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, YeastLow, BioGrid |
| CAD | 790 | RPSA | 3921 | pp | -- | int.I2D: IntAct_Yeast |
| PSMB2 | 5690 | PSMC1 | 5700 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium |
| PSMA2 | 5683 | PSMA3 | 5684 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct, IntAct_Fly, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastHigh, BIND_Yeast, HPRD; int.HPRD: yeast 2-hybrid |
| PSMA1 | 5682 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium |
| ACO2 | 50 | DDX18 | 8886 | pp | -- | int.I2D: YeastLow |
| ACTB | 60 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| RUVBL2 | 10856 | PHB2 | 11331 | pd | > | reg.ITFP.txt: no annot |
| RUVBL2 | 10856 | PHB2 | 11331 | pp | -- | int.I2D: IntAct_Yeast |
| CAD | 790 | SMARCA4 | 6597 | pd | < | reg.ITFP.txt: no annot |
| PSMB2 | 5690 | PSMD1 | 5707 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA2 | 5683 | PSMB2 | 5690 | pp | -- | int.Intact: MI:0914(association); int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct, INTEROLOG, MINT_Yeast, YeastHigh, IntAct_Yeast, Krogan_Core |
| PSMC1 | 5700 | PHB2 | 11331 | pp | -- | int.I2D: BioGrid_Yeast |
| ENO1 | 2023 | PSMA3 | 5684 | pp | -- | int.I2D: BioGrid |
Related GO terms (654)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 1.58e-20 | 2.59e-16 | 5.920 | 13 | 23 | 73 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 3.35e-20 | 5.46e-16 | 5.843 | 13 | 23 | 77 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 3.39e-19 | 5.53e-15 | 5.972 | 12 | 24 | 65 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 5.30e-19 | 8.66e-15 | 5.555 | 13 | 23 | 94 |
| GO:0016032 | viral process | 5.42e-19 | 8.84e-15 | 3.725 | 21 | 55 | 540 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 8.86e-19 | 1.45e-14 | 5.865 | 12 | 24 | 70 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 1.42e-18 | 2.32e-14 | 6.225 | 11 | 21 | 50 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1.81e-18 | 2.96e-14 | 5.785 | 12 | 24 | 74 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 4.19e-18 | 6.84e-14 | 5.691 | 12 | 25 | 79 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 6.10e-18 | 9.96e-14 | 4.988 | 14 | 33 | 150 |
| GO:0000502 | proteasome complex | 8.54e-18 | 1.39e-13 | 6.011 | 11 | 22 | 58 |
| GO:0005654 | nucleoplasm | 2.48e-17 | 4.05e-13 | 2.957 | 25 | 83 | 1095 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 3.31e-17 | 5.40e-13 | 5.847 | 11 | 22 | 65 |
| GO:0010467 | gene expression | 4.26e-17 | 6.95e-13 | 3.416 | 21 | 58 | 669 |
| GO:0016071 | mRNA metabolic process | 4.94e-17 | 8.06e-13 | 4.515 | 15 | 34 | 223 |
| GO:0016070 | RNA metabolic process | 2.29e-16 | 3.74e-12 | 4.368 | 15 | 34 | 247 |
| GO:0005829 | cytosol | 2.30e-16 | 3.76e-12 | 2.131 | 33 | 125 | 2562 |
| GO:0016020 | membrane | 1.21e-14 | 1.98e-10 | 2.395 | 27 | 80 | 1746 |
| GO:0000278 | mitotic cell cycle | 1.28e-14 | 2.09e-10 | 3.773 | 16 | 52 | 398 |
| GO:0042981 | regulation of apoptotic process | 1.32e-14 | 2.16e-10 | 4.756 | 12 | 26 | 151 |
| GO:0000209 | protein polyubiquitination | 2.61e-14 | 4.26e-10 | 5.011 | 11 | 21 | 116 |
| GO:0034641 | cellular nitrogen compound metabolic process | 9.02e-14 | 1.47e-09 | 4.527 | 12 | 25 | 177 |
| GO:0070062 | extracellular vesicular exosome | 1.55e-13 | 2.54e-09 | 2.019 | 30 | 98 | 2516 |
| GO:0022624 | proteasome accessory complex | 5.65e-12 | 9.22e-08 | 6.907 | 6 | 9 | 17 |
| GO:0043066 | negative regulation of apoptotic process | 1.47e-11 | 2.40e-07 | 3.459 | 14 | 30 | 433 |
| GO:0005838 | proteasome regulatory particle | 1.38e-10 | 2.26e-06 | 7.146 | 5 | 7 | 12 |
| GO:0005839 | proteasome core complex | 1.48e-09 | 2.41e-05 | 6.561 | 5 | 11 | 18 |
| GO:0004298 | threonine-type endopeptidase activity | 2.66e-09 | 4.35e-05 | 6.409 | 5 | 11 | 20 |
| GO:0044281 | small molecule metabolic process | 1.01e-08 | 1.65e-04 | 2.241 | 18 | 57 | 1295 |
| GO:0005730 | nucleolus | 1.63e-08 | 2.66e-04 | 2.014 | 20 | 70 | 1684 |
| GO:0005634 | nucleus | 2.31e-08 | 3.76e-04 | 1.217 | 33 | 131 | 4828 |
| GO:0005515 | protein binding | 2.65e-08 | 4.32e-04 | 1.038 | 37 | 172 | 6127 |
| GO:0044822 | poly(A) RNA binding | 3.54e-08 | 5.78e-04 | 2.335 | 16 | 50 | 1078 |
| GO:0030957 | Tat protein binding | 4.75e-07 | 7.74e-03 | 7.409 | 3 | 4 | 6 |
| GO:0006915 | apoptotic process | 6.08e-07 | 9.93e-03 | 2.712 | 11 | 34 | 571 |
| GO:0030529 | ribonucleoprotein complex | 9.83e-07 | 1.60e-02 | 4.162 | 6 | 8 | 114 |
| GO:0019773 | proteasome core complex, alpha-subunit complex | 1.32e-06 | 2.16e-02 | 6.994 | 3 | 5 | 8 |
| GO:0006414 | translational elongation | 7.60e-06 | 1.24e-01 | 4.192 | 5 | 11 | 93 |
| GO:0051082 | unfolded protein binding | 8.44e-06 | 1.38e-01 | 4.162 | 5 | 6 | 95 |
| GO:0035267 | NuA4 histone acetyltransferase complex | 8.49e-06 | 1.39e-01 | 6.187 | 3 | 4 | 14 |
| GO:0005925 | focal adhesion | 1.11e-05 | 1.80e-01 | 2.878 | 8 | 18 | 370 |
| GO:0006281 | DNA repair | 1.12e-05 | 1.82e-01 | 3.172 | 7 | 22 | 264 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 1.87e-05 | 3.05e-01 | 4.655 | 4 | 5 | 54 |
| GO:0019058 | viral life cycle | 2.14e-05 | 3.49e-01 | 3.886 | 5 | 10 | 115 |
| GO:0003678 | DNA helicase activity | 2.24e-05 | 3.65e-01 | 5.747 | 3 | 3 | 19 |
| GO:0043234 | protein complex | 2.55e-05 | 4.16e-01 | 2.988 | 7 | 17 | 300 |
| GO:0043044 | ATP-dependent chromatin remodeling | 4.06e-05 | 6.62e-01 | 5.471 | 3 | 4 | 23 |
| GO:0005844 | polysome | 5.25e-05 | 8.56e-01 | 5.351 | 3 | 4 | 25 |
| GO:0016887 | ATPase activity | 6.29e-05 | 1.00e+00 | 3.561 | 5 | 7 | 144 |
| GO:0031492 | nucleosomal DNA binding | 7.43e-05 | 1.00e+00 | 5.187 | 3 | 4 | 28 |
| GO:0019083 | viral transcription | 9.28e-05 | 1.00e+00 | 4.070 | 4 | 8 | 81 |
| GO:0006271 | DNA strand elongation involved in DNA replication | 1.01e-04 | 1.00e+00 | 5.040 | 3 | 9 | 31 |
| GO:0006415 | translational termination | 1.23e-04 | 1.00e+00 | 3.967 | 4 | 8 | 87 |
| GO:0016363 | nuclear matrix | 1.52e-04 | 1.00e+00 | 3.886 | 4 | 11 | 92 |
| GO:0001649 | osteoblast differentiation | 1.72e-04 | 1.00e+00 | 3.840 | 4 | 6 | 95 |
| GO:0050681 | androgen receptor binding | 1.87e-04 | 1.00e+00 | 4.747 | 3 | 4 | 38 |
| GO:0022627 | cytosolic small ribosomal subunit | 2.03e-04 | 1.00e+00 | 4.709 | 3 | 3 | 39 |
| GO:0000812 | Swr1 complex | 2.34e-04 | 1.00e+00 | 6.409 | 2 | 3 | 8 |
| GO:0070182 | DNA polymerase binding | 2.34e-04 | 1.00e+00 | 6.409 | 2 | 2 | 8 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 2.44e-04 | 1.00e+00 | 3.709 | 4 | 8 | 104 |
| GO:0006200 | ATP catabolic process | 2.48e-04 | 1.00e+00 | 2.751 | 6 | 14 | 303 |
| GO:0032508 | DNA duplex unwinding | 2.53e-04 | 1.00e+00 | 4.602 | 3 | 4 | 42 |
| GO:0006283 | transcription-coupled nucleotide-excision repair | 3.32e-04 | 1.00e+00 | 4.471 | 3 | 8 | 46 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 3.47e-04 | 1.00e+00 | 3.577 | 4 | 10 | 114 |
| GO:0005524 | ATP binding | 4.23e-04 | 1.00e+00 | 1.591 | 12 | 46 | 1354 |
| GO:0005813 | centrosome | 4.52e-04 | 1.00e+00 | 2.589 | 6 | 12 | 339 |
| GO:0043968 | histone H2A acetylation | 5.49e-04 | 1.00e+00 | 5.825 | 2 | 3 | 12 |
| GO:0044267 | cellular protein metabolic process | 5.67e-04 | 1.00e+00 | 2.266 | 7 | 24 | 495 |
| GO:0003723 | RNA binding | 5.76e-04 | 1.00e+00 | 2.523 | 6 | 19 | 355 |
| GO:0006413 | translational initiation | 5.87e-04 | 1.00e+00 | 3.376 | 4 | 12 | 131 |
| GO:0006412 | translation | 6.13e-04 | 1.00e+00 | 2.855 | 5 | 15 | 235 |
| GO:0030234 | enzyme regulator activity | 6.47e-04 | 1.00e+00 | 5.709 | 2 | 3 | 13 |
| GO:0051087 | chaperone binding | 6.91e-04 | 1.00e+00 | 4.112 | 3 | 6 | 59 |
| GO:0031011 | Ino80 complex | 7.54e-04 | 1.00e+00 | 5.602 | 2 | 3 | 14 |
| GO:0003735 | structural constituent of ribosome | 7.73e-04 | 1.00e+00 | 3.270 | 4 | 8 | 141 |
| GO:0042026 | protein refolding | 8.68e-04 | 1.00e+00 | 5.503 | 2 | 2 | 15 |
| GO:0042176 | regulation of protein catabolic process | 9.90e-04 | 1.00e+00 | 5.409 | 2 | 3 | 16 |
| GO:0006289 | nucleotide-excision repair | 1.09e-03 | 1.00e+00 | 3.886 | 3 | 12 | 69 |
| GO:0017025 | TBP-class protein binding | 1.26e-03 | 1.00e+00 | 5.240 | 2 | 2 | 18 |
| GO:0005719 | nuclear euchromatin | 1.56e-03 | 1.00e+00 | 5.088 | 2 | 2 | 20 |
| GO:0006298 | mismatch repair | 1.56e-03 | 1.00e+00 | 5.088 | 2 | 6 | 20 |
| GO:0004860 | protein kinase inhibitor activity | 1.72e-03 | 1.00e+00 | 5.017 | 2 | 2 | 21 |
| GO:0033574 | response to testosterone | 1.88e-03 | 1.00e+00 | 4.950 | 2 | 2 | 22 |
| GO:0006297 | nucleotide-excision repair, DNA gap filling | 1.88e-03 | 1.00e+00 | 4.950 | 2 | 5 | 22 |
| GO:0032201 | telomere maintenance via semi-conservative replication | 1.88e-03 | 1.00e+00 | 4.950 | 2 | 7 | 22 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.88e-03 | 1.00e+00 | 4.950 | 2 | 4 | 22 |
| GO:0031625 | ubiquitin protein ligase binding | 1.91e-03 | 1.00e+00 | 2.918 | 4 | 13 | 180 |
| GO:0019901 | protein kinase binding | 2.41e-03 | 1.00e+00 | 2.409 | 5 | 21 | 320 |
| GO:0006611 | protein export from nucleus | 2.43e-03 | 1.00e+00 | 4.766 | 2 | 4 | 25 |
| GO:0042470 | melanosome | 2.50e-03 | 1.00e+00 | 3.471 | 3 | 10 | 92 |
| GO:0000722 | telomere maintenance via recombination | 2.63e-03 | 1.00e+00 | 4.709 | 2 | 7 | 26 |
| GO:0071339 | MLL1 complex | 2.84e-03 | 1.00e+00 | 4.655 | 2 | 3 | 27 |
| GO:0034080 | CENP-A containing nucleosome assembly | 2.84e-03 | 1.00e+00 | 4.655 | 2 | 2 | 27 |
| GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0002502 | peptide antigen assembly with MHC class I protein complex | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0032077 | positive regulation of deoxyribonuclease activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0000701 | purine-specific mismatch base pair DNA N-glycosylase activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0048291 | isotype switching to IgG isotypes | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0052066 | entry of symbiont into host cell by promotion of host phagocytosis | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0060699 | regulation of endoribonuclease activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0019521 | D-gluconate metabolic process | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0016074 | snoRNA metabolic process | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0002368 | B cell cytokine production | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0060735 | regulation of eIF2 alpha phosphorylation by dsRNA | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0070335 | aspartate binding | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:1990259 | histone-glutamine methyltransferase activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0043626 | PCNA complex | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:2000425 | regulation of apoptotic cell clearance | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:1990258 | histone glutamine methylation | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0004151 | dihydroorotase activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0004070 | aspartate carbamoyltransferase activity | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:2000536 | negative regulation of entry of bacterium into host cell | 2.94e-03 | 1.00e+00 | 8.409 | 1 | 1 | 1 |
| GO:0003924 | GTPase activity | 2.95e-03 | 1.00e+00 | 2.744 | 4 | 9 | 203 |
| GO:0043967 | histone H4 acetylation | 3.05e-03 | 1.00e+00 | 4.602 | 2 | 3 | 28 |
| GO:0006099 | tricarboxylic acid cycle | 3.27e-03 | 1.00e+00 | 4.551 | 2 | 3 | 29 |
| GO:0006184 | GTP catabolic process | 3.93e-03 | 1.00e+00 | 2.628 | 4 | 9 | 220 |
| GO:0033572 | transferrin transport | 3.97e-03 | 1.00e+00 | 4.409 | 2 | 6 | 32 |
| GO:0007067 | mitotic nuclear division | 4.67e-03 | 1.00e+00 | 2.558 | 4 | 13 | 231 |
| GO:0005759 | mitochondrial matrix | 4.82e-03 | 1.00e+00 | 2.545 | 4 | 12 | 233 |
| GO:0003713 | transcription coactivator activity | 5.27e-03 | 1.00e+00 | 2.509 | 4 | 10 | 239 |
| GO:0006325 | chromatin organization | 5.64e-03 | 1.00e+00 | 3.052 | 3 | 4 | 123 |
| GO:0006096 | glycolytic process | 5.86e-03 | 1.00e+00 | 4.124 | 2 | 4 | 39 |
| GO:0032092 | positive regulation of protein binding | 5.86e-03 | 1.00e+00 | 4.124 | 2 | 3 | 39 |
| GO:0006284 | base-excision repair | 5.86e-03 | 1.00e+00 | 4.124 | 2 | 7 | 39 |
| GO:0006272 | leading strand elongation | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 2 | 2 |
| GO:0045252 | oxoglutarate dehydrogenase complex | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 2 | 2 |
| GO:0034686 | integrin alphav-beta8 complex | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0035887 | aortic smooth muscle cell differentiation | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0003994 | aconitate hydratase activity | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0032071 | regulation of endodeoxyribonuclease activity | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0006407 | rRNA export from nucleus | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0042824 | MHC class I peptide loading complex | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0001846 | opsonin binding | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:1990430 | extracellular matrix protein binding | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0061034 | olfactory bulb mitral cell layer development | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0050748 | negative regulation of lipoprotein metabolic process | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0019322 | pentose biosynthetic process | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0034683 | integrin alphav-beta3 complex | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0030337 | DNA polymerase processivity factor activity | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0051538 | 3 iron, 4 sulfur cluster binding | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0070557 | PCNA-p21 complex | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0005055 | laminin receptor activity | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0034684 | integrin alphav-beta5 complex | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0007127 | meiosis I | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0007070 | negative regulation of transcription from RNA polymerase II promoter during mitosis | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 2 | 2 |
| GO:0070409 | carbamoyl phosphate biosynthetic process | 5.87e-03 | 1.00e+00 | 7.409 | 1 | 1 | 2 |
| GO:0009615 | response to virus | 6.86e-03 | 1.00e+00 | 2.950 | 3 | 6 | 132 |
| GO:0000790 | nuclear chromatin | 7.00e-03 | 1.00e+00 | 2.939 | 3 | 7 | 133 |
| GO:0015030 | Cajal body | 7.74e-03 | 1.00e+00 | 3.918 | 2 | 2 | 45 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 7.91e-03 | 1.00e+00 | 2.003 | 5 | 14 | 424 |
| GO:0045727 | positive regulation of translation | 8.07e-03 | 1.00e+00 | 3.886 | 2 | 4 | 46 |
| GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0010424 | DNA methylation on cytosine within a CG sequence | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:1900126 | negative regulation of hyaluronan biosynthetic process | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0006458 | 'de novo' protein folding | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0007403 | glial cell fate determination | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0009051 | pentose-phosphate shunt, oxidative branch | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0030135 | coated vesicle | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0030953 | astral microtubule organization | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0005726 | perichromatin fibrils | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0044205 | 'de novo' UMP biosynthetic process | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0032139 | dinucleotide insertion or deletion binding | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0071899 | negative regulation of estrogen receptor binding | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0071733 | transcriptional activation by promoter-enhancer looping | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0001832 | blastocyst growth | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0055106 | ubiquitin-protein transferase regulator activity | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0000056 | ribosomal small subunit export from nucleus | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0071439 | clathrin complex | 8.80e-03 | 1.00e+00 | 6.825 | 1 | 1 | 3 |
| GO:0006457 | protein folding | 9.55e-03 | 1.00e+00 | 2.775 | 3 | 8 | 149 |
| GO:0006091 | generation of precursor metabolites and energy | 9.85e-03 | 1.00e+00 | 3.737 | 2 | 3 | 51 |
| GO:0040008 | regulation of growth | 9.85e-03 | 1.00e+00 | 3.737 | 2 | 3 | 51 |
| GO:0003684 | damaged DNA binding | 9.85e-03 | 1.00e+00 | 3.737 | 2 | 11 | 51 |
| GO:0006986 | response to unfolded protein | 9.85e-03 | 1.00e+00 | 3.737 | 2 | 2 | 51 |
| GO:0003725 | double-stranded RNA binding | 1.10e-02 | 1.00e+00 | 3.655 | 2 | 6 | 54 |
| GO:0002039 | p53 binding | 1.14e-02 | 1.00e+00 | 3.628 | 2 | 7 | 55 |
| GO:0030686 | 90S preribosome | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0019788 | NEDD8 ligase activity | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0001652 | granular component | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0031428 | box C/D snoRNP complex | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0043141 | ATP-dependent 5'-3' DNA helicase activity | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0007098 | centrosome cycle | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:2000510 | positive regulation of dendritic cell chemotaxis | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0000212 | meiotic spindle organization | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0032051 | clathrin light chain binding | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0010826 | negative regulation of centrosome duplication | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0000015 | phosphopyruvate hydratase complex | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 2 | 4 |
| GO:0004634 | phosphopyruvate hydratase activity | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 2 | 4 |
| GO:0060318 | definitive erythrocyte differentiation | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0006104 | succinyl-CoA metabolic process | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0006543 | glutamine catabolic process | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0001835 | blastocyst hatching | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 2 | 4 |
| GO:0031467 | Cul7-RING ubiquitin ligase complex | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0000055 | ribosomal large subunit export from nucleus | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0032369 | negative regulation of lipid transport | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0051208 | sequestering of calcium ion | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:1903077 | negative regulation of protein localization to plasma membrane | 1.17e-02 | 1.00e+00 | 6.409 | 1 | 1 | 4 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.26e-02 | 1.00e+00 | 2.628 | 3 | 12 | 165 |
| GO:0000723 | telomere maintenance | 1.30e-02 | 1.00e+00 | 3.527 | 2 | 8 | 59 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.32e-02 | 1.00e+00 | 2.602 | 3 | 5 | 168 |
| GO:0005737 | cytoplasm | 1.39e-02 | 1.00e+00 | 0.700 | 19 | 98 | 3976 |
| GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0005672 | transcription factor TFIIA complex | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:2000001 | regulation of DNA damage checkpoint | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0048562 | embryonic organ morphogenesis | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0043248 | proteasome assembly | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0030891 | VCB complex | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 2 | 5 |
| GO:0005827 | polar microtubule | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0043405 | regulation of MAP kinase activity | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0006734 | NADH metabolic process | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0003407 | neural retina development | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0051414 | response to cortisol | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 2 | 5 |
| GO:0038027 | apolipoprotein A-I-mediated signaling pathway | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0046599 | regulation of centriole replication | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0031461 | cullin-RING ubiquitin ligase complex | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0071169 | establishment of protein localization to chromatin | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0046696 | lipopolysaccharide receptor complex | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0006102 | isocitrate metabolic process | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0042255 | ribosome assembly | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0000730 | DNA recombinase assembly | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 3 | 5 |
| GO:0048730 | epidermis morphogenesis | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0030976 | thiamine pyrophosphate binding | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 5 |
| GO:0003714 | transcription corepressor activity | 1.56e-02 | 1.00e+00 | 2.511 | 3 | 7 | 179 |
| GO:0005525 | GTP binding | 1.56e-02 | 1.00e+00 | 2.052 | 4 | 11 | 328 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 1.66e-02 | 1.00e+00 | 3.343 | 2 | 6 | 67 |
| GO:0006310 | DNA recombination | 1.66e-02 | 1.00e+00 | 3.343 | 2 | 4 | 67 |
| GO:0006338 | chromatin remodeling | 1.71e-02 | 1.00e+00 | 3.322 | 2 | 4 | 68 |
| GO:0003688 | DNA replication origin binding | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0005663 | DNA replication factor C complex | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0006101 | citrate metabolic process | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0030118 | clathrin coat | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0003697 | single-stranded DNA binding | 1.75e-02 | 1.00e+00 | 3.301 | 2 | 9 | 69 |
| GO:0021695 | cerebellar cortex development | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0046134 | pyrimidine nucleoside biosynthetic process | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0050764 | regulation of phagocytosis | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0040020 | regulation of meiosis | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0002181 | cytoplasmic translation | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0030130 | clathrin coat of trans-Golgi network vesicle | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0060744 | mammary gland branching involved in thelarche | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0006346 | methylation-dependent chromatin silencing | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0021860 | pyramidal neuron development | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0043023 | ribosomal large subunit binding | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 2 | 6 |
| GO:0033993 | response to lipid | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 2 | 6 |
| GO:0031466 | Cul5-RING ubiquitin ligase complex | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 1 | 6 |
| GO:0032405 | MutLalpha complex binding | 1.75e-02 | 1.00e+00 | 5.825 | 1 | 2 | 6 |
| GO:0042921 | glucocorticoid receptor signaling pathway | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 1 | 7 |
| GO:0001849 | complement component C1q binding | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 1 | 7 |
| GO:0010888 | negative regulation of lipid storage | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 2 | 7 |
| GO:0031462 | Cul2-RING ubiquitin ligase complex | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 2 | 7 |
| GO:0031994 | insulin-like growth factor I binding | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 1 | 7 |
| GO:0000028 | ribosomal small subunit assembly | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 1 | 7 |
| GO:0033180 | proton-transporting V-type ATPase, V1 domain | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 2 | 7 |
| GO:0030132 | clathrin coat of coated pit | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 1 | 7 |
| GO:0000930 | gamma-tubulin complex | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 1 | 7 |
| GO:0006334 | nucleosome assembly | 2.26e-02 | 1.00e+00 | 3.106 | 2 | 4 | 79 |
| GO:0071013 | catalytic step 2 spliceosome | 2.26e-02 | 1.00e+00 | 3.106 | 2 | 7 | 79 |
| GO:0006554 | lysine catabolic process | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 2 | 8 |
| GO:0031616 | spindle pole centrosome | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 1 | 8 |
| GO:0045116 | protein neddylation | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 2 | 8 |
| GO:0043596 | nuclear replication fork | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 1 | 8 |
| GO:0033018 | sarcoplasmic reticulum lumen | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 1 | 8 |
| GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 2 | 8 |
| GO:0001055 | RNA polymerase II activity | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 3 | 8 |
| GO:0070688 | MLL5-L complex | 2.33e-02 | 1.00e+00 | 5.409 | 1 | 1 | 8 |
| GO:0001889 | liver development | 2.42e-02 | 1.00e+00 | 3.052 | 2 | 3 | 82 |
| GO:0005681 | spliceosomal complex | 2.48e-02 | 1.00e+00 | 3.034 | 2 | 3 | 83 |
| GO:0044183 | protein binding involved in protein folding | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0045717 | negative regulation of fatty acid biosynthetic process | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0033690 | positive regulation of osteoblast proliferation | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0014075 | response to amine | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0042555 | MCM complex | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 2 | 9 |
| GO:0008494 | translation activator activity | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0006228 | UTP biosynthetic process | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0031000 | response to caffeine | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 2 | 9 |
| GO:0022417 | protein maturation by protein folding | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0022027 | interkinetic nuclear migration | 2.62e-02 | 1.00e+00 | 5.240 | 1 | 1 | 9 |
| GO:0047485 | protein N-terminus binding | 2.65e-02 | 1.00e+00 | 2.983 | 2 | 4 | 86 |
| GO:0002199 | zona pellucida receptor complex | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 10 |
| GO:0043032 | positive regulation of macrophage activation | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 10 |
| GO:0070307 | lens fiber cell development | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 2 | 10 |
| GO:0051604 | protein maturation | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 10 |
| GO:0043024 | ribosomal small subunit binding | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 10 |
| GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 10 |
| GO:0021756 | striatum development | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 10 |
| GO:0050821 | protein stabilization | 2.94e-02 | 1.00e+00 | 2.902 | 2 | 2 | 91 |
| GO:0005200 | structural constituent of cytoskeleton | 3.06e-02 | 1.00e+00 | 2.870 | 2 | 7 | 93 |
| GO:0008380 | RNA splicing | 3.07e-02 | 1.00e+00 | 2.136 | 3 | 13 | 232 |
| GO:0032727 | positive regulation of interferon-alpha production | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 1 | 11 |
| GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 1 | 11 |
| GO:0045120 | pronucleus | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 1 | 11 |
| GO:0006098 | pentose-phosphate shunt | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 3 | 11 |
| GO:0010569 | regulation of double-strand break repair via homologous recombination | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 1 | 11 |
| GO:0031571 | mitotic G1 DNA damage checkpoint | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 3 | 11 |
| GO:0001054 | RNA polymerase I activity | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 3 | 11 |
| GO:0043923 | positive regulation by host of viral transcription | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 2 | 11 |
| GO:0071564 | npBAF complex | 3.19e-02 | 1.00e+00 | 4.950 | 1 | 2 | 11 |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 2 | 12 |
| GO:0005736 | DNA-directed RNA polymerase I complex | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 3 | 12 |
| GO:0021794 | thalamus development | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 1 | 12 |
| GO:0019985 | translesion synthesis | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 2 | 12 |
| GO:0032886 | regulation of microtubule-based process | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 4 | 12 |
| GO:0071565 | nBAF complex | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 2 | 12 |
| GO:0006275 | regulation of DNA replication | 3.47e-02 | 1.00e+00 | 4.825 | 1 | 2 | 12 |
| GO:0001530 | lipopolysaccharide binding | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 2 | 13 |
| GO:0042994 | cytoplasmic sequestering of transcription factor | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 1 | 13 |
| GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 2 | 13 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 1 | 13 |
| GO:0005662 | DNA replication factor A complex | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 3 | 13 |
| GO:0008266 | poly(U) RNA binding | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 1 | 13 |
| GO:0051131 | chaperone-mediated protein complex assembly | 3.76e-02 | 1.00e+00 | 4.709 | 1 | 1 | 13 |
| GO:0043277 | apoptotic cell clearance | 4.04e-02 | 1.00e+00 | 4.602 | 1 | 1 | 14 |
| GO:0071285 | cellular response to lithium ion | 4.04e-02 | 1.00e+00 | 4.602 | 1 | 2 | 14 |
| GO:0007020 | microtubule nucleation | 4.04e-02 | 1.00e+00 | 4.602 | 1 | 1 | 14 |
| GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 15 |
| GO:0046961 | proton-transporting ATPase activity, rotational mechanism | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 3 | 15 |
| GO:0016514 | SWI/SNF complex | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 3 | 15 |
| GO:0060749 | mammary gland alveolus development | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 15 |
| GO:0035066 | positive regulation of histone acetylation | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 15 |
| GO:0050431 | transforming growth factor beta binding | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 15 |
| GO:0060347 | heart trabecula formation | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 15 |
| GO:0030308 | negative regulation of cell growth | 4.36e-02 | 1.00e+00 | 2.589 | 2 | 6 | 113 |
| GO:0005819 | spindle | 4.43e-02 | 1.00e+00 | 2.577 | 2 | 7 | 114 |
| GO:0072562 | blood microparticle | 4.57e-02 | 1.00e+00 | 2.551 | 2 | 4 | 116 |
| GO:0042562 | hormone binding | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0030902 | hindbrain development | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0050998 | nitric-oxide synthase binding | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0001056 | RNA polymerase III activity | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 3 | 16 |
| GO:0005665 | DNA-directed RNA polymerase II, core complex | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 4 | 16 |
| GO:0046034 | ATP metabolic process | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0071682 | endocytic vesicle lumen | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0006103 | 2-oxoglutarate metabolic process | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0031589 | cell-substrate adhesion | 4.61e-02 | 1.00e+00 | 4.409 | 1 | 1 | 16 |
| GO:0044237 | cellular metabolic process | 4.71e-02 | 1.00e+00 | 2.527 | 2 | 3 | 118 |
| GO:0006337 | nucleosome disassembly | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 4 | 17 |
| GO:0003746 | translation elongation factor activity | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 3 | 17 |
| GO:0031528 | microvillus membrane | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 17 |
| GO:0031258 | lamellipodium membrane | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 2 | 17 |
| GO:0075733 | intracellular transport of virus | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 2 | 17 |
| GO:0070577 | lysine-acetylated histone binding | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 17 |
| GO:0031527 | filopodium membrane | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 17 |
| GO:0010243 | response to organonitrogen compound | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 2 | 17 |
| GO:0050919 | negative chemotaxis | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 17 |
| GO:0005666 | DNA-directed RNA polymerase III complex | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 3 | 17 |
| GO:0050870 | positive regulation of T cell activation | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 17 |
| GO:0046718 | viral entry into host cell | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 1 | 18 |
| GO:0071392 | cellular response to estradiol stimulus | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 1 | 18 |
| GO:0031122 | cytoplasmic microtubule organization | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 2 | 18 |
| GO:0035861 | site of double-strand break | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 1 | 18 |
| GO:0004004 | ATP-dependent RNA helicase activity | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 2 | 18 |
| GO:0006386 | termination of RNA polymerase III transcription | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 3 | 18 |
| GO:0006541 | glutamine metabolic process | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 1 | 18 |
| GO:0006385 | transcription elongation from RNA polymerase III promoter | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 3 | 18 |
| GO:0070371 | ERK1 and ERK2 cascade | 5.17e-02 | 1.00e+00 | 4.240 | 1 | 1 | 18 |
| GO:0006260 | DNA replication | 5.22e-02 | 1.00e+00 | 2.444 | 2 | 12 | 125 |
| GO:0005506 | iron ion binding | 5.37e-02 | 1.00e+00 | 2.421 | 2 | 3 | 127 |
| GO:0006259 | DNA metabolic process | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 3 | 19 |
| GO:0050840 | extracellular matrix binding | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0032733 | positive regulation of interleukin-10 production | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0007088 | regulation of mitosis | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0055007 | cardiac muscle cell differentiation | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0034113 | heterotypic cell-cell adhesion | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0048863 | stem cell differentiation | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0031430 | M band | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0030866 | cortical actin cytoskeleton organization | 5.45e-02 | 1.00e+00 | 4.162 | 1 | 1 | 19 |
| GO:0042802 | identical protein binding | 5.57e-02 | 1.00e+00 | 1.470 | 4 | 18 | 491 |
| GO:0015078 | hydrogen ion transmembrane transporter activity | 5.72e-02 | 1.00e+00 | 4.088 | 1 | 3 | 20 |
| GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 5.72e-02 | 1.00e+00 | 4.088 | 1 | 2 | 20 |
| GO:0090398 | cellular senescence | 5.72e-02 | 1.00e+00 | 4.088 | 1 | 1 | 20 |
| GO:0005680 | anaphase-promoting complex | 5.72e-02 | 1.00e+00 | 4.088 | 1 | 4 | 20 |
| GO:0071364 | cellular response to epidermal growth factor stimulus | 6.00e-02 | 1.00e+00 | 4.017 | 1 | 1 | 21 |
| GO:0000718 | nucleotide-excision repair, DNA damage removal | 6.00e-02 | 1.00e+00 | 4.017 | 1 | 5 | 21 |
| GO:0046686 | response to cadmium ion | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 3 | 22 |
| GO:0005790 | smooth endoplasmic reticulum | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 1 | 22 |
| GO:0000792 | heterochromatin | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 2 | 22 |
| GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 3 | 22 |
| GO:0030863 | cortical cytoskeleton | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 1 | 22 |
| GO:0006270 | DNA replication initiation | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 5 | 22 |
| GO:0007052 | mitotic spindle organization | 6.28e-02 | 1.00e+00 | 3.950 | 1 | 2 | 22 |
| GO:0007507 | heart development | 6.46e-02 | 1.00e+00 | 2.270 | 2 | 5 | 141 |
| GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 6.55e-02 | 1.00e+00 | 3.886 | 1 | 2 | 23 |
| GO:0031463 | Cul3-RING ubiquitin ligase complex | 6.55e-02 | 1.00e+00 | 3.886 | 1 | 2 | 23 |
| GO:0043236 | laminin binding | 6.55e-02 | 1.00e+00 | 3.886 | 1 | 1 | 23 |
| GO:0045787 | positive regulation of cell cycle | 6.55e-02 | 1.00e+00 | 3.886 | 1 | 1 | 23 |
| GO:0008305 | integrin complex | 6.55e-02 | 1.00e+00 | 3.886 | 1 | 1 | 23 |
| GO:0006513 | protein monoubiquitination | 6.55e-02 | 1.00e+00 | 3.886 | 1 | 1 | 23 |
| GO:0008286 | insulin receptor signaling pathway | 6.70e-02 | 1.00e+00 | 2.240 | 2 | 6 | 144 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 1 | 24 |
| GO:0001105 | RNA polymerase II transcription coactivator activity | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 1 | 24 |
| GO:0050766 | positive regulation of phagocytosis | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 1 | 24 |
| GO:0000794 | condensed nuclear chromosome | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 2 | 24 |
| GO:0006206 | pyrimidine nucleobase metabolic process | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 2 | 24 |
| GO:0043388 | positive regulation of DNA binding | 6.83e-02 | 1.00e+00 | 3.825 | 1 | 1 | 24 |
| GO:0061024 | membrane organization | 6.86e-02 | 1.00e+00 | 2.220 | 2 | 5 | 146 |
| GO:0042100 | B cell proliferation | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 1 | 25 |
| GO:0001968 | fibronectin binding | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 1 | 25 |
| GO:0051059 | NF-kappaB binding | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 3 | 25 |
| GO:0022008 | neurogenesis | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 1 | 25 |
| GO:0007569 | cell aging | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 2 | 25 |
| GO:0042113 | B cell activation | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 2 | 25 |
| GO:0017144 | drug metabolic process | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 1 | 25 |
| GO:0032735 | positive regulation of interleukin-12 production | 7.10e-02 | 1.00e+00 | 3.766 | 1 | 1 | 25 |
| GO:0010628 | positive regulation of gene expression | 7.11e-02 | 1.00e+00 | 2.190 | 2 | 4 | 149 |
| GO:0070979 | protein K11-linked ubiquitination | 7.38e-02 | 1.00e+00 | 3.709 | 1 | 3 | 26 |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 7.38e-02 | 1.00e+00 | 3.709 | 1 | 2 | 26 |
| GO:0035987 | endodermal cell differentiation | 7.38e-02 | 1.00e+00 | 3.709 | 1 | 1 | 26 |
| GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane | 7.38e-02 | 1.00e+00 | 3.709 | 1 | 1 | 26 |
| GO:0004003 | ATP-dependent DNA helicase activity | 7.65e-02 | 1.00e+00 | 3.655 | 1 | 3 | 27 |
| GO:0019843 | rRNA binding | 7.65e-02 | 1.00e+00 | 3.655 | 1 | 3 | 27 |
| GO:0006913 | nucleocytoplasmic transport | 7.65e-02 | 1.00e+00 | 3.655 | 1 | 2 | 27 |
| GO:0030331 | estrogen receptor binding | 7.65e-02 | 1.00e+00 | 3.655 | 1 | 2 | 27 |
| GO:0007339 | binding of sperm to zona pellucida | 7.65e-02 | 1.00e+00 | 3.655 | 1 | 1 | 27 |
| GO:0015991 | ATP hydrolysis coupled proton transport | 7.92e-02 | 1.00e+00 | 3.602 | 1 | 4 | 28 |
| GO:0043022 | ribosome binding | 7.92e-02 | 1.00e+00 | 3.602 | 1 | 3 | 28 |
| GO:0019894 | kinesin binding | 7.92e-02 | 1.00e+00 | 3.602 | 1 | 1 | 28 |
| GO:0030177 | positive regulation of Wnt signaling pathway | 7.92e-02 | 1.00e+00 | 3.602 | 1 | 3 | 28 |
| GO:0019005 | SCF ubiquitin ligase complex | 8.19e-02 | 1.00e+00 | 3.551 | 1 | 1 | 29 |
| GO:0003730 | mRNA 3'-UTR binding | 8.19e-02 | 1.00e+00 | 3.551 | 1 | 2 | 29 |
| GO:0030669 | clathrin-coated endocytic vesicle membrane | 8.19e-02 | 1.00e+00 | 3.551 | 1 | 1 | 29 |
| GO:0001618 | virus receptor activity | 8.46e-02 | 1.00e+00 | 3.503 | 1 | 1 | 30 |
| GO:0006360 | transcription from RNA polymerase I promoter | 8.46e-02 | 1.00e+00 | 3.503 | 1 | 4 | 30 |
| GO:0006370 | 7-methylguanosine mRNA capping | 8.46e-02 | 1.00e+00 | 3.503 | 1 | 4 | 30 |
| GO:0034504 | protein localization to nucleus | 8.46e-02 | 1.00e+00 | 3.503 | 1 | 2 | 30 |
| GO:0035116 | embryonic hindlimb morphogenesis | 8.46e-02 | 1.00e+00 | 3.503 | 1 | 1 | 30 |
| GO:0061077 | chaperone-mediated protein folding | 8.73e-02 | 1.00e+00 | 3.455 | 1 | 2 | 31 |
| GO:0031623 | receptor internalization | 8.73e-02 | 1.00e+00 | 3.455 | 1 | 1 | 31 |
| GO:0010827 | regulation of glucose transport | 8.73e-02 | 1.00e+00 | 3.455 | 1 | 1 | 31 |
| GO:0007094 | mitotic spindle assembly checkpoint | 8.73e-02 | 1.00e+00 | 3.455 | 1 | 5 | 31 |
| GO:0050661 | NADP binding | 9.00e-02 | 1.00e+00 | 3.409 | 1 | 1 | 32 |
| GO:0034644 | cellular response to UV | 9.00e-02 | 1.00e+00 | 3.409 | 1 | 5 | 32 |
| GO:0015992 | proton transport | 9.00e-02 | 1.00e+00 | 3.409 | 1 | 3 | 32 |
| GO:0006886 | intracellular protein transport | 9.18e-02 | 1.00e+00 | 1.975 | 2 | 4 | 173 |
| GO:0045335 | phagocytic vesicle | 9.27e-02 | 1.00e+00 | 3.365 | 1 | 2 | 33 |
| GO:0030971 | receptor tyrosine kinase binding | 9.27e-02 | 1.00e+00 | 3.365 | 1 | 2 | 33 |
| GO:0031072 | heat shock protein binding | 9.27e-02 | 1.00e+00 | 3.365 | 1 | 2 | 33 |
| GO:0008094 | DNA-dependent ATPase activity | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 1 | 34 |
| GO:0008180 | COP9 signalosome | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 1 | 34 |
| GO:0051701 | interaction with host | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 4 | 34 |
| GO:0005876 | spindle microtubule | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 3 | 34 |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 9.80e-02 | 1.00e+00 | 3.280 | 1 | 1 | 35 |
| GO:0042277 | peptide binding | 9.80e-02 | 1.00e+00 | 3.280 | 1 | 2 | 35 |
| GO:0045599 | negative regulation of fat cell differentiation | 9.80e-02 | 1.00e+00 | 3.280 | 1 | 2 | 35 |
| GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 9.80e-02 | 1.00e+00 | 3.280 | 1 | 2 | 35 |
| GO:0019904 | protein domain specific binding | 9.91e-02 | 1.00e+00 | 1.910 | 2 | 6 | 181 |
| GO:0009897 | external side of plasma membrane | 1.01e-01 | 1.00e+00 | 1.894 | 2 | 4 | 183 |
| GO:0032588 | trans-Golgi network membrane | 1.01e-01 | 1.00e+00 | 3.240 | 1 | 1 | 36 |
| GO:0034332 | adherens junction organization | 1.01e-01 | 1.00e+00 | 3.240 | 1 | 1 | 36 |
| GO:0034446 | substrate adhesion-dependent cell spreading | 1.01e-01 | 1.00e+00 | 3.240 | 1 | 2 | 36 |
| GO:0032755 | positive regulation of interleukin-6 production | 1.01e-01 | 1.00e+00 | 3.240 | 1 | 2 | 36 |
| GO:0001895 | retina homeostasis | 1.01e-01 | 1.00e+00 | 3.240 | 1 | 1 | 36 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.02e-01 | 1.00e+00 | 1.886 | 2 | 8 | 184 |
| GO:0016301 | kinase activity | 1.03e-01 | 1.00e+00 | 3.200 | 1 | 3 | 37 |
| GO:0051084 | 'de novo' posttranslational protein folding | 1.03e-01 | 1.00e+00 | 3.200 | 1 | 4 | 37 |
| GO:0005245 | voltage-gated calcium channel activity | 1.03e-01 | 1.00e+00 | 3.200 | 1 | 1 | 37 |
| GO:0018107 | peptidyl-threonine phosphorylation | 1.03e-01 | 1.00e+00 | 3.200 | 1 | 1 | 37 |
| GO:0051539 | 4 iron, 4 sulfur cluster binding | 1.03e-01 | 1.00e+00 | 3.200 | 1 | 3 | 37 |
| GO:0070527 | platelet aggregation | 1.06e-01 | 1.00e+00 | 3.162 | 1 | 2 | 38 |
| GO:0045740 | positive regulation of DNA replication | 1.06e-01 | 1.00e+00 | 3.162 | 1 | 2 | 38 |
| GO:0090382 | phagosome maturation | 1.06e-01 | 1.00e+00 | 3.162 | 1 | 5 | 38 |
| GO:0032729 | positive regulation of interferon-gamma production | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 2 | 39 |
| GO:0021766 | hippocampus development | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 4 | 39 |
| GO:0008033 | tRNA processing | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 1 | 39 |
| GO:0006383 | transcription from RNA polymerase III promoter | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 3 | 39 |
| GO:0031490 | chromatin DNA binding | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 2 | 39 |
| GO:0007595 | lactation | 1.09e-01 | 1.00e+00 | 3.124 | 1 | 2 | 39 |
| GO:0000781 | chromosome, telomeric region | 1.11e-01 | 1.00e+00 | 3.088 | 1 | 2 | 40 |
| GO:0017148 | negative regulation of translation | 1.14e-01 | 1.00e+00 | 3.052 | 1 | 1 | 41 |
| GO:0030521 | androgen receptor signaling pathway | 1.14e-01 | 1.00e+00 | 3.052 | 1 | 2 | 41 |
| GO:0045785 | positive regulation of cell adhesion | 1.14e-01 | 1.00e+00 | 3.052 | 1 | 5 | 41 |
| GO:0043195 | terminal bouton | 1.14e-01 | 1.00e+00 | 3.052 | 1 | 1 | 41 |
| GO:0051259 | protein oligomerization | 1.14e-01 | 1.00e+00 | 3.052 | 1 | 2 | 41 |
| GO:0021987 | cerebral cortex development | 1.16e-01 | 1.00e+00 | 3.017 | 1 | 3 | 42 |
| GO:0005902 | microvillus | 1.16e-01 | 1.00e+00 | 3.017 | 1 | 2 | 42 |
| GO:0042110 | T cell activation | 1.19e-01 | 1.00e+00 | 2.983 | 1 | 3 | 43 |
| GO:0014070 | response to organic cyclic compound | 1.19e-01 | 1.00e+00 | 2.983 | 1 | 3 | 43 |
| GO:0007286 | spermatid development | 1.22e-01 | 1.00e+00 | 2.950 | 1 | 1 | 44 |
| GO:0005080 | protein kinase C binding | 1.22e-01 | 1.00e+00 | 2.950 | 1 | 1 | 44 |
| GO:0006892 | post-Golgi vesicle-mediated transport | 1.22e-01 | 1.00e+00 | 2.950 | 1 | 2 | 44 |
| GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding | 1.22e-01 | 1.00e+00 | 2.950 | 1 | 2 | 44 |
| GO:0050434 | positive regulation of viral transcription | 1.22e-01 | 1.00e+00 | 2.950 | 1 | 5 | 44 |
| GO:0006094 | gluconeogenesis | 1.24e-01 | 1.00e+00 | 2.918 | 1 | 3 | 45 |
| GO:0043966 | histone H3 acetylation | 1.24e-01 | 1.00e+00 | 2.918 | 1 | 2 | 45 |
| GO:0030136 | clathrin-coated vesicle | 1.27e-01 | 1.00e+00 | 2.886 | 1 | 1 | 46 |
| GO:0044297 | cell body | 1.27e-01 | 1.00e+00 | 2.886 | 1 | 2 | 46 |
| GO:0021762 | substantia nigra development | 1.27e-01 | 1.00e+00 | 2.886 | 1 | 1 | 46 |
| GO:0045665 | negative regulation of neuron differentiation | 1.27e-01 | 1.00e+00 | 2.886 | 1 | 2 | 46 |
| GO:0009986 | cell surface | 1.27e-01 | 1.00e+00 | 1.273 | 3 | 9 | 422 |
| GO:0030216 | keratinocyte differentiation | 1.29e-01 | 1.00e+00 | 2.855 | 1 | 1 | 47 |
| GO:0019827 | stem cell maintenance | 1.32e-01 | 1.00e+00 | 2.825 | 1 | 1 | 48 |
| GO:0006950 | response to stress | 1.32e-01 | 1.00e+00 | 2.825 | 1 | 3 | 48 |
| GO:0019003 | GDP binding | 1.32e-01 | 1.00e+00 | 2.825 | 1 | 2 | 48 |
| GO:0022625 | cytosolic large ribosomal subunit | 1.35e-01 | 1.00e+00 | 2.795 | 1 | 5 | 49 |
| GO:0031100 | organ regeneration | 1.37e-01 | 1.00e+00 | 2.766 | 1 | 4 | 50 |
| GO:0001948 | glycoprotein binding | 1.37e-01 | 1.00e+00 | 2.766 | 1 | 3 | 50 |
| GO:0035690 | cellular response to drug | 1.37e-01 | 1.00e+00 | 2.766 | 1 | 2 | 50 |
| GO:0016049 | cell growth | 1.37e-01 | 1.00e+00 | 2.766 | 1 | 1 | 50 |
| GO:0005905 | coated pit | 1.40e-01 | 1.00e+00 | 2.737 | 1 | 2 | 51 |
| GO:0001669 | acrosomal vesicle | 1.40e-01 | 1.00e+00 | 2.737 | 1 | 1 | 51 |
| GO:0000902 | cell morphogenesis | 1.40e-01 | 1.00e+00 | 2.737 | 1 | 2 | 51 |
| GO:0030900 | forebrain development | 1.40e-01 | 1.00e+00 | 2.737 | 1 | 1 | 51 |
| GO:0034976 | response to endoplasmic reticulum stress | 1.42e-01 | 1.00e+00 | 2.709 | 1 | 1 | 52 |
| GO:0005622 | intracellular | 1.43e-01 | 1.00e+00 | 1.589 | 2 | 5 | 226 |
| GO:0000226 | microtubule cytoskeleton organization | 1.50e-01 | 1.00e+00 | 2.628 | 1 | 3 | 55 |
| GO:0004386 | helicase activity | 1.52e-01 | 1.00e+00 | 2.602 | 1 | 4 | 56 |
| GO:0000932 | cytoplasmic mRNA processing body | 1.52e-01 | 1.00e+00 | 2.602 | 1 | 3 | 56 |
| GO:0008104 | protein localization | 1.52e-01 | 1.00e+00 | 2.602 | 1 | 3 | 56 |
| GO:0006879 | cellular iron ion homeostasis | 1.55e-01 | 1.00e+00 | 2.577 | 1 | 5 | 57 |
| GO:0000724 | double-strand break repair via homologous recombination | 1.55e-01 | 1.00e+00 | 2.577 | 1 | 6 | 57 |
| GO:0012505 | endomembrane system | 1.55e-01 | 1.00e+00 | 2.577 | 1 | 2 | 57 |
| GO:0030097 | hemopoiesis | 1.57e-01 | 1.00e+00 | 2.551 | 1 | 3 | 58 |
| GO:0002244 | hematopoietic progenitor cell differentiation | 1.57e-01 | 1.00e+00 | 2.551 | 1 | 1 | 58 |
| GO:0045216 | cell-cell junction organization | 1.60e-01 | 1.00e+00 | 2.527 | 1 | 2 | 59 |
| GO:0005643 | nuclear pore | 1.60e-01 | 1.00e+00 | 2.527 | 1 | 4 | 59 |
| GO:0005840 | ribosome | 1.60e-01 | 1.00e+00 | 2.527 | 1 | 2 | 59 |
| GO:0001570 | vasculogenesis | 1.60e-01 | 1.00e+00 | 2.527 | 1 | 1 | 59 |
| GO:0031966 | mitochondrial membrane | 1.60e-01 | 1.00e+00 | 2.527 | 1 | 1 | 59 |
| GO:0032481 | positive regulation of type I interferon production | 1.65e-01 | 1.00e+00 | 2.479 | 1 | 6 | 61 |
| GO:0006302 | double-strand break repair | 1.67e-01 | 1.00e+00 | 2.455 | 1 | 8 | 62 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 1.67e-01 | 1.00e+00 | 2.455 | 1 | 2 | 62 |
| GO:0019903 | protein phosphatase binding | 1.70e-01 | 1.00e+00 | 2.432 | 1 | 4 | 63 |
| GO:0042995 | cell projection | 1.70e-01 | 1.00e+00 | 2.432 | 1 | 6 | 63 |
| GO:0032869 | cellular response to insulin stimulus | 1.72e-01 | 1.00e+00 | 2.409 | 1 | 3 | 64 |
| GO:0005615 | extracellular space | 1.74e-01 | 1.00e+00 | 0.751 | 5 | 17 | 1010 |
| GO:0030855 | epithelial cell differentiation | 1.75e-01 | 1.00e+00 | 2.387 | 1 | 4 | 65 |
| GO:0006469 | negative regulation of protein kinase activity | 1.75e-01 | 1.00e+00 | 2.387 | 1 | 2 | 65 |
| GO:0030141 | secretory granule | 1.79e-01 | 1.00e+00 | 2.343 | 1 | 2 | 67 |
| GO:0034329 | cell junction assembly | 1.89e-01 | 1.00e+00 | 2.260 | 1 | 1 | 71 |
| GO:0042393 | histone binding | 1.89e-01 | 1.00e+00 | 2.260 | 1 | 3 | 71 |
| GO:0032587 | ruffle membrane | 1.91e-01 | 1.00e+00 | 2.240 | 1 | 4 | 72 |
| GO:0005975 | carbohydrate metabolic process | 1.93e-01 | 1.00e+00 | 1.311 | 2 | 5 | 274 |
| GO:0003729 | mRNA binding | 1.94e-01 | 1.00e+00 | 2.220 | 1 | 4 | 73 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 1.94e-01 | 1.00e+00 | 1.306 | 2 | 6 | 275 |
| GO:0032355 | response to estradiol | 1.94e-01 | 1.00e+00 | 2.220 | 1 | 5 | 73 |
| GO:0000785 | chromatin | 1.94e-01 | 1.00e+00 | 2.220 | 1 | 5 | 73 |
| GO:0055086 | nucleobase-containing small molecule metabolic process | 1.94e-01 | 1.00e+00 | 2.220 | 1 | 5 | 73 |
| GO:0007283 | spermatogenesis | 1.95e-01 | 1.00e+00 | 1.301 | 2 | 6 | 276 |
| GO:0002020 | protease binding | 1.96e-01 | 1.00e+00 | 2.200 | 1 | 4 | 74 |
| GO:0006874 | cellular calcium ion homeostasis | 2.01e-01 | 1.00e+00 | 2.162 | 1 | 1 | 76 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 2.01e-01 | 1.00e+00 | 2.162 | 1 | 3 | 76 |
| GO:0003677 | DNA binding | 2.03e-01 | 1.00e+00 | 0.595 | 6 | 26 | 1351 |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 2.03e-01 | 1.00e+00 | 2.143 | 1 | 5 | 77 |
| GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 2.03e-01 | 1.00e+00 | 2.143 | 1 | 6 | 77 |
| GO:0007229 | integrin-mediated signaling pathway | 2.06e-01 | 1.00e+00 | 2.124 | 1 | 2 | 78 |
| GO:0019899 | enzyme binding | 2.08e-01 | 1.00e+00 | 1.240 | 2 | 11 | 288 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 2.10e-01 | 1.00e+00 | 2.088 | 1 | 2 | 80 |
| GO:0007565 | female pregnancy | 2.10e-01 | 1.00e+00 | 2.088 | 1 | 2 | 80 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 2.13e-01 | 1.00e+00 | 2.070 | 1 | 2 | 81 |
| GO:0001726 | ruffle | 2.15e-01 | 1.00e+00 | 2.052 | 1 | 4 | 82 |
| GO:0030198 | extracellular matrix organization | 2.15e-01 | 1.00e+00 | 1.205 | 2 | 3 | 295 |
| GO:0005743 | mitochondrial inner membrane | 2.21e-01 | 1.00e+00 | 1.181 | 2 | 5 | 300 |
| GO:0006898 | receptor-mediated endocytosis | 2.24e-01 | 1.00e+00 | 1.983 | 1 | 2 | 86 |
| GO:0007160 | cell-matrix adhesion | 2.29e-01 | 1.00e+00 | 1.950 | 1 | 3 | 88 |
| GO:0018279 | protein N-linked glycosylation via asparagine | 2.36e-01 | 1.00e+00 | 1.902 | 1 | 2 | 91 |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 2.38e-01 | 1.00e+00 | 1.886 | 1 | 4 | 92 |
| GO:0006928 | cellular component movement | 2.38e-01 | 1.00e+00 | 1.886 | 1 | 7 | 92 |
| GO:0016605 | PML body | 2.38e-01 | 1.00e+00 | 1.886 | 1 | 5 | 92 |
| GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity | 2.40e-01 | 1.00e+00 | 1.870 | 1 | 5 | 93 |
| GO:0006364 | rRNA processing | 2.47e-01 | 1.00e+00 | 1.825 | 1 | 5 | 96 |
| GO:0005178 | integrin binding | 2.49e-01 | 1.00e+00 | 1.810 | 1 | 2 | 97 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.50e-01 | 1.00e+00 | 0.792 | 3 | 12 | 589 |
| GO:0007411 | axon guidance | 2.50e-01 | 1.00e+00 | 1.056 | 2 | 9 | 327 |
| GO:0071456 | cellular response to hypoxia | 2.51e-01 | 1.00e+00 | 1.795 | 1 | 4 | 98 |
| GO:0008283 | cell proliferation | 2.54e-01 | 1.00e+00 | 1.039 | 2 | 12 | 331 |
| GO:0043231 | intracellular membrane-bounded organelle | 2.55e-01 | 1.00e+00 | 1.034 | 2 | 8 | 332 |
| GO:0051726 | regulation of cell cycle | 2.60e-01 | 1.00e+00 | 1.737 | 1 | 3 | 102 |
| GO:0070588 | calcium ion transmembrane transport | 2.69e-01 | 1.00e+00 | 1.682 | 1 | 1 | 106 |
| GO:0014069 | postsynaptic density | 2.69e-01 | 1.00e+00 | 1.682 | 1 | 1 | 106 |
| GO:0045087 | innate immune response | 2.71e-01 | 1.00e+00 | 0.728 | 3 | 20 | 616 |
| GO:0005815 | microtubule organizing center | 2.78e-01 | 1.00e+00 | 1.628 | 1 | 4 | 110 |
| GO:0050900 | leukocyte migration | 2.80e-01 | 1.00e+00 | 1.615 | 1 | 1 | 111 |
| GO:0015630 | microtubule cytoskeleton | 2.82e-01 | 1.00e+00 | 1.602 | 1 | 5 | 112 |
| GO:0048015 | phosphatidylinositol-mediated signaling | 2.88e-01 | 1.00e+00 | 1.564 | 1 | 3 | 115 |
| GO:0006006 | glucose metabolic process | 2.97e-01 | 1.00e+00 | 1.515 | 1 | 4 | 119 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 3.09e-01 | 1.00e+00 | 1.444 | 1 | 4 | 125 |
| GO:0007219 | Notch signaling pathway | 3.09e-01 | 1.00e+00 | 1.444 | 1 | 4 | 125 |
| GO:0007050 | cell cycle arrest | 3.11e-01 | 1.00e+00 | 1.432 | 1 | 7 | 126 |
| GO:0007155 | cell adhesion | 3.12e-01 | 1.00e+00 | 0.825 | 2 | 8 | 384 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 3.13e-01 | 1.00e+00 | 1.421 | 1 | 5 | 127 |
| GO:0030335 | positive regulation of cell migration | 3.19e-01 | 1.00e+00 | 1.387 | 1 | 6 | 130 |
| GO:0008284 | positive regulation of cell proliferation | 3.21e-01 | 1.00e+00 | 0.795 | 2 | 8 | 392 |
| GO:0016477 | cell migration | 3.21e-01 | 1.00e+00 | 1.376 | 1 | 6 | 131 |
| GO:0006351 | transcription, DNA-templated | 3.22e-01 | 1.00e+00 | 0.364 | 6 | 25 | 1585 |
| GO:0031982 | vesicle | 3.27e-01 | 1.00e+00 | 1.343 | 1 | 10 | 134 |
| GO:0046982 | protein heterodimerization activity | 3.29e-01 | 1.00e+00 | 0.769 | 2 | 11 | 399 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 3.33e-01 | 1.00e+00 | 1.311 | 1 | 7 | 137 |
| GO:0044255 | cellular lipid metabolic process | 3.39e-01 | 1.00e+00 | 1.280 | 1 | 4 | 140 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.57e-01 | 1.00e+00 | 0.678 | 2 | 12 | 425 |
| GO:0001666 | response to hypoxia | 3.58e-01 | 1.00e+00 | 1.181 | 1 | 2 | 150 |
| GO:0030246 | carbohydrate binding | 3.60e-01 | 1.00e+00 | 1.171 | 1 | 1 | 151 |
| GO:0005739 | mitochondrion | 3.70e-01 | 1.00e+00 | 0.379 | 4 | 24 | 1046 |
| GO:0005788 | endoplasmic reticulum lumen | 3.72e-01 | 1.00e+00 | 1.115 | 1 | 1 | 157 |
| GO:0005769 | early endosome | 3.74e-01 | 1.00e+00 | 1.106 | 1 | 2 | 158 |
| GO:0046777 | protein autophosphorylation | 3.74e-01 | 1.00e+00 | 1.106 | 1 | 3 | 158 |
| GO:0005198 | structural molecule activity | 3.75e-01 | 1.00e+00 | 1.097 | 1 | 4 | 159 |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | 3.75e-01 | 1.00e+00 | 1.097 | 1 | 4 | 159 |
| GO:0006397 | mRNA processing | 3.94e-01 | 1.00e+00 | 1.009 | 1 | 3 | 169 |
| GO:0038095 | Fc-epsilon receptor signaling pathway | 3.97e-01 | 1.00e+00 | 0.992 | 1 | 7 | 171 |
| GO:0007596 | blood coagulation | 3.98e-01 | 1.00e+00 | 0.551 | 2 | 14 | 464 |
| GO:0030424 | axon | 3.99e-01 | 1.00e+00 | 0.983 | 1 | 3 | 172 |
| GO:0000287 | magnesium ion binding | 4.03e-01 | 1.00e+00 | 0.967 | 1 | 5 | 174 |
| GO:0016607 | nuclear speck | 4.04e-01 | 1.00e+00 | 0.958 | 1 | 4 | 175 |
| GO:0004672 | protein kinase activity | 4.10e-01 | 1.00e+00 | 0.934 | 1 | 2 | 178 |
| GO:0043687 | post-translational protein modification | 4.15e-01 | 1.00e+00 | 0.910 | 1 | 4 | 181 |
| GO:0005578 | proteinaceous extracellular matrix | 4.18e-01 | 1.00e+00 | 0.894 | 1 | 1 | 183 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 4.22e-01 | 1.00e+00 | 0.482 | 2 | 17 | 487 |
| GO:0032403 | protein complex binding | 4.22e-01 | 1.00e+00 | 0.878 | 1 | 7 | 185 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 4.32e-01 | 1.00e+00 | 0.832 | 1 | 4 | 191 |
| GO:0001525 | angiogenesis | 4.47e-01 | 1.00e+00 | 0.766 | 1 | 4 | 200 |
| GO:0005765 | lysosomal membrane | 4.76e-01 | 1.00e+00 | 0.641 | 1 | 5 | 218 |
| GO:0008134 | transcription factor binding | 5.18e-01 | 1.00e+00 | 0.467 | 1 | 8 | 246 |
| GO:0043025 | neuronal cell body | 5.30e-01 | 1.00e+00 | 0.421 | 1 | 4 | 254 |
| GO:0004842 | ubiquitin-protein transferase activity | 5.32e-01 | 1.00e+00 | 0.409 | 1 | 4 | 256 |
| GO:0000166 | nucleotide binding | 5.54e-01 | 1.00e+00 | 0.322 | 1 | 6 | 272 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 5.56e-01 | 1.00e+00 | 0.317 | 1 | 6 | 273 |
| GO:0043065 | positive regulation of apoptotic process | 5.57e-01 | 1.00e+00 | 0.311 | 1 | 8 | 274 |
| GO:0042493 | response to drug | 5.75e-01 | 1.00e+00 | 0.240 | 1 | 11 | 288 |
| GO:0005794 | Golgi apparatus | 5.75e-01 | 1.00e+00 | 0.065 | 2 | 14 | 650 |
| GO:0007264 | small GTPase mediated signal transduction | 5.78e-01 | 1.00e+00 | 0.230 | 1 | 3 | 290 |
| GO:0016567 | protein ubiquitination | 5.89e-01 | 1.00e+00 | 0.185 | 1 | 5 | 299 |
| GO:0005856 | cytoskeleton | 6.03e-01 | 1.00e+00 | 0.129 | 1 | 8 | 311 |
| GO:0008270 | zinc ion binding | 6.16e-01 | 1.00e+00 | -0.065 | 3 | 12 | 1067 |
| GO:0030154 | cell differentiation | 6.20e-01 | 1.00e+00 | 0.065 | 1 | 5 | 325 |
| GO:0003682 | chromatin binding | 6.30e-01 | 1.00e+00 | 0.026 | 1 | 12 | 334 |
| GO:0006355 | regulation of transcription, DNA-templated | 6.39e-01 | 1.00e+00 | -0.114 | 3 | 17 | 1104 |
| GO:0008285 | negative regulation of cell proliferation | 6.65e-01 | 1.00e+00 | -0.110 | 1 | 11 | 367 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 6.96e-01 | 1.00e+00 | -0.254 | 2 | 19 | 811 |
| GO:0006508 | proteolysis | 7.06e-01 | 1.00e+00 | -0.270 | 1 | 9 | 410 |
| GO:0005886 | plasma membrane | 7.51e-01 | 1.00e+00 | -0.252 | 7 | 38 | 2834 |
| GO:0006468 | protein phosphorylation | 7.52e-01 | 1.00e+00 | -0.458 | 1 | 10 | 467 |
| GO:0055114 | oxidation-reduction process | 7.63e-01 | 1.00e+00 | -0.500 | 1 | 11 | 481 |
| GO:0007165 | signal transduction | 7.77e-01 | 1.00e+00 | -0.482 | 2 | 17 | 950 |
| GO:0055085 | transmembrane transport | 7.85e-01 | 1.00e+00 | -0.596 | 1 | 8 | 514 |
| GO:0048471 | perinuclear region of cytoplasm | 7.91e-01 | 1.00e+00 | -0.621 | 1 | 12 | 523 |
| GO:0005509 | calcium ion binding | 8.29e-01 | 1.00e+00 | -0.793 | 1 | 8 | 589 |
| GO:0005783 | endoplasmic reticulum | 8.40e-01 | 1.00e+00 | -0.843 | 1 | 9 | 610 |
| GO:0042803 | protein homodimerization activity | 8.43e-01 | 1.00e+00 | -0.860 | 1 | 11 | 617 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 8.95e-01 | 1.00e+00 | -1.137 | 1 | 11 | 748 |
| GO:0046872 | metal ion binding | 9.37e-01 | 1.00e+00 | -1.107 | 2 | 24 | 1465 |
| GO:0005887 | integral component of plasma membrane | 9.46e-01 | 1.00e+00 | -1.499 | 1 | 7 | 961 |
| GO:0005576 | extracellular region | 9.59e-01 | 1.00e+00 | -1.625 | 1 | 9 | 1049 |
| GO:0016021 | integral component of membrane | 1.00e+00 | 1.00e+00 | -2.868 | 1 | 15 | 2483 |