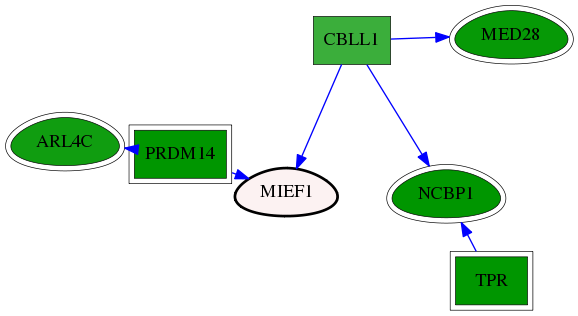
| GO:0034972 | histone H3-R26 methylation | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:1901673 | regulation of spindle assembly involved in mitosis | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:0043578 | nuclear matrix organization | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:0010965 | regulation of mitotic sister chromatid separation | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:0001827 | inner cell mass cell fate commitment | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:0006404 | RNA import into nucleus | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:0031453 | positive regulation of heterochromatin assembly | 7.17e-04 | 1.00e+00 | 10.446 | 1 | 1 | 1 |
| GO:0000189 | MAPK import into nucleus | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 2 |
| GO:0031990 | mRNA export from nucleus in response to heat stress | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 2 |
| GO:0042306 | regulation of protein import into nucleus | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 2 |
| GO:0005846 | nuclear cap binding complex | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 2 |
| GO:0070840 | dynein complex binding | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 2 |
| GO:0046832 | negative regulation of RNA export from nucleus | 1.43e-03 | 1.00e+00 | 9.446 | 1 | 1 | 2 |
| GO:0006405 | RNA export from nucleus | 2.15e-03 | 1.00e+00 | 8.861 | 1 | 1 | 3 |
| GO:0010793 | regulation of mRNA export from nucleus | 2.15e-03 | 1.00e+00 | 8.861 | 1 | 1 | 3 |
| GO:0005515 | protein binding | 2.39e-03 | 1.00e+00 | 1.244 | 7 | 198 | 4124 |
| GO:0051151 | negative regulation of smooth muscle cell differentiation | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0045292 | mRNA cis splicing, via spliceosome | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0044030 | regulation of DNA methylation | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0030718 | germ-line stem cell maintenance | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0031442 | positive regulation of mRNA 3'-end processing | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0006999 | nuclear pore organization | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0070849 | response to epidermal growth factor | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0044615 | nuclear pore nuclear basket | 2.86e-03 | 1.00e+00 | 8.446 | 1 | 1 | 4 |
| GO:0046825 | regulation of protein export from nucleus | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 5 |
| GO:0043495 | protein anchor | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 5 |
| GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 6 |
| GO:0090141 | positive regulation of mitochondrial fission | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 6 |
| GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 6 |
| GO:0000339 | RNA cap binding | 5.01e-03 | 1.00e+00 | 7.639 | 1 | 1 | 7 |
| GO:0005868 | cytoplasmic dynein complex | 5.01e-03 | 1.00e+00 | 7.639 | 1 | 1 | 7 |
| GO:0005845 | mRNA cap binding complex | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 8 |
| GO:0042405 | nuclear inclusion body | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 8 |
| GO:0051292 | nuclear pore complex assembly | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 4 | 8 |
| GO:0035457 | cellular response to interferon-alpha | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 8 |
| GO:0032456 | endocytic recycling | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 2 | 8 |
| GO:0090314 | positive regulation of protein targeting to membrane | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 2 | 9 |
| GO:0006379 | mRNA cleavage | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 2 | 9 |
| GO:0045947 | negative regulation of translational initiation | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 1 | 9 |
| GO:0008053 | mitochondrial fusion | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 10 |
| GO:0040029 | regulation of gene expression, epigenetic | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 10 |
| GO:0051019 | mitogen-activated protein kinase binding | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 11 |
| GO:0008334 | histone mRNA metabolic process | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 11 |
| GO:0090316 | positive regulation of intracellular protein transport | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 11 |
| GO:0045807 | positive regulation of endocytosis | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 11 |
| GO:0005487 | nucleocytoplasmic transporter activity | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 2 | 11 |
| GO:0000266 | mitochondrial fission | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 2 | 11 |
| GO:0046827 | positive regulation of protein export from nucleus | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 11 |
| GO:0042307 | positive regulation of protein import into nucleus | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 3 | 12 |
| GO:0034399 | nuclear periphery | 8.57e-03 | 1.00e+00 | 6.861 | 1 | 3 | 12 |
| GO:0048873 | homeostasis of number of cells within a tissue | 1.00e-02 | 1.00e+00 | 6.639 | 1 | 2 | 14 |
| GO:0043014 | alpha-tubulin binding | 1.07e-02 | 1.00e+00 | 6.539 | 1 | 1 | 15 |
| GO:0001708 | cell fate specification | 1.07e-02 | 1.00e+00 | 6.539 | 1 | 1 | 15 |
| GO:0030864 | cortical actin cytoskeleton | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 16 |
| GO:0006611 | protein export from nucleus | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 18 |
| GO:0031047 | gene silencing by RNA | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 2 | 18 |
| GO:0009566 | fertilization | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 18 |
| GO:0043531 | ADP binding | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 2 | 20 |
| GO:0015631 | tubulin binding | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 20 |
| GO:0072686 | mitotic spindle | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 2 | 20 |
| GO:0034660 | ncRNA metabolic process | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 20 |
| GO:0031647 | regulation of protein stability | 1.50e-02 | 1.00e+00 | 6.054 | 1 | 2 | 21 |
| GO:0006606 | protein import into nucleus | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 3 | 22 |
| GO:0000387 | spliceosomal snRNP assembly | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 22 |
| GO:0034605 | cellular response to heat | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 22 |
| GO:0007281 | germ cell development | 1.57e-02 | 1.00e+00 | 5.987 | 1 | 1 | 22 |
| GO:0007162 | negative regulation of cell adhesion | 1.71e-02 | 1.00e+00 | 5.861 | 1 | 2 | 24 |
| GO:0006370 | 7-methylguanosine mRNA capping | 1.71e-02 | 1.00e+00 | 5.861 | 1 | 5 | 24 |
| GO:0016592 | mediator complex | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 6 | 25 |
| GO:0010827 | regulation of glucose transport | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 7 | 25 |
| GO:0007094 | mitotic spindle assembly checkpoint | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 4 | 25 |
| GO:0031072 | heat shock protein binding | 1.99e-02 | 1.00e+00 | 5.639 | 1 | 1 | 28 |
| GO:0006446 | regulation of translational initiation | 2.06e-02 | 1.00e+00 | 5.588 | 1 | 2 | 29 |
| GO:0019898 | extrinsic component of membrane | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 1 | 30 |
| GO:0008168 | methyltransferase activity | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 2 | 30 |
| GO:0007077 | mitotic nuclear envelope disassembly | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 9 | 32 |
| GO:0019003 | GDP binding | 2.41e-02 | 1.00e+00 | 5.359 | 1 | 2 | 34 |
| GO:0042802 | identical protein binding | 2.44e-02 | 1.00e+00 | 2.978 | 2 | 15 | 354 |
| GO:0000902 | cell morphogenesis | 2.48e-02 | 1.00e+00 | 5.317 | 1 | 5 | 35 |
| GO:0050434 | positive regulation of viral transcription | 2.48e-02 | 1.00e+00 | 5.317 | 1 | 5 | 35 |
| GO:0031124 | mRNA 3'-end processing | 2.48e-02 | 1.00e+00 | 5.317 | 1 | 4 | 35 |
| GO:0008645 | hexose transport | 2.55e-02 | 1.00e+00 | 5.276 | 1 | 8 | 36 |
| GO:0030175 | filopodium | 2.62e-02 | 1.00e+00 | 5.237 | 1 | 1 | 37 |
| GO:0006369 | termination of RNA polymerase II transcription | 2.69e-02 | 1.00e+00 | 5.198 | 1 | 5 | 38 |
| GO:0019827 | stem cell maintenance | 2.76e-02 | 1.00e+00 | 5.161 | 1 | 4 | 39 |
| GO:0006406 | mRNA export from nucleus | 3.11e-02 | 1.00e+00 | 4.987 | 1 | 7 | 44 |
| GO:0015758 | glucose transport | 3.39e-02 | 1.00e+00 | 4.861 | 1 | 8 | 48 |
| GO:0000151 | ubiquitin ligase complex | 3.39e-02 | 1.00e+00 | 4.861 | 1 | 4 | 48 |
| GO:0016032 | viral process | 3.48e-02 | 1.00e+00 | 2.705 | 2 | 30 | 428 |
| GO:0005777 | peroxisome | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 2 | 50 |
| GO:0005643 | nuclear pore | 3.60e-02 | 1.00e+00 | 4.774 | 1 | 12 | 51 |
| GO:0006368 | transcription elongation from RNA polymerase II promoter | 3.67e-02 | 1.00e+00 | 4.746 | 1 | 8 | 52 |
| GO:0000776 | kinetochore | 3.88e-02 | 1.00e+00 | 4.665 | 1 | 8 | 55 |
| GO:0003729 | mRNA binding | 4.02e-02 | 1.00e+00 | 4.613 | 1 | 4 | 57 |
| GO:0016337 | single organismal cell-cell adhesion | 4.36e-02 | 1.00e+00 | 4.492 | 1 | 3 | 62 |
| GO:0005741 | mitochondrial outer membrane | 5.53e-02 | 1.00e+00 | 4.142 | 1 | 2 | 79 |
| GO:0030529 | ribonucleoprotein complex | 5.73e-02 | 1.00e+00 | 4.088 | 1 | 5 | 82 |
| GO:0005215 | transporter activity | 5.73e-02 | 1.00e+00 | 4.088 | 1 | 5 | 82 |
| GO:0005635 | nuclear envelope | 5.80e-02 | 1.00e+00 | 4.071 | 1 | 6 | 83 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 5.87e-02 | 1.00e+00 | 4.054 | 1 | 5 | 84 |
| GO:0030335 | positive regulation of cell migration | 7.02e-02 | 1.00e+00 | 3.788 | 1 | 2 | 101 |
| GO:0005739 | mitochondrion | 7.61e-02 | 1.00e+00 | 2.082 | 2 | 32 | 659 |
| GO:0003924 | GTPase activity | 8.23e-02 | 1.00e+00 | 3.551 | 1 | 7 | 119 |
| GO:0006355 | regulation of transcription, DNA-templated | 8.79e-02 | 1.00e+00 | 1.964 | 2 | 31 | 715 |
| GO:0000398 | mRNA splicing, via spliceosome | 8.83e-02 | 1.00e+00 | 3.446 | 1 | 15 | 128 |
| GO:0031965 | nuclear membrane | 8.96e-02 | 1.00e+00 | 3.424 | 1 | 8 | 130 |
| GO:0006184 | GTP catabolic process | 8.96e-02 | 1.00e+00 | 3.424 | 1 | 7 | 130 |
| GO:0016874 | ligase activity | 1.00e-01 | 1.00e+00 | 3.256 | 1 | 11 | 146 |
| GO:0044822 | poly(A) RNA binding | 1.07e-01 | 1.00e+00 | 1.804 | 2 | 45 | 799 |
| GO:0008380 | RNA splicing | 1.12e-01 | 1.00e+00 | 3.080 | 1 | 21 | 165 |
| GO:0007067 | mitotic nuclear division | 1.12e-01 | 1.00e+00 | 3.080 | 1 | 9 | 165 |
| GO:0016071 | mRNA metabolic process | 1.16e-01 | 1.00e+00 | 3.037 | 1 | 10 | 170 |
| GO:0019221 | cytokine-mediated signaling pathway | 1.17e-01 | 1.00e+00 | 3.020 | 1 | 10 | 172 |
| GO:0007264 | small GTPase mediated signal transduction | 1.18e-01 | 1.00e+00 | 3.003 | 1 | 8 | 174 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.22e-01 | 1.00e+00 | 2.962 | 1 | 17 | 179 |
| GO:0003779 | actin binding | 1.22e-01 | 1.00e+00 | 2.962 | 1 | 3 | 179 |
| GO:0005654 | nucleoplasm | 1.25e-01 | 1.00e+00 | 1.671 | 2 | 68 | 876 |
| GO:0005975 | carbohydrate metabolic process | 1.26e-01 | 1.00e+00 | 2.907 | 1 | 10 | 186 |
| GO:0016567 | protein ubiquitination | 1.27e-01 | 1.00e+00 | 2.891 | 1 | 14 | 188 |
| GO:0016070 | RNA metabolic process | 1.27e-01 | 1.00e+00 | 2.891 | 1 | 10 | 188 |
| GO:0005525 | GTP binding | 1.33e-01 | 1.00e+00 | 2.824 | 1 | 12 | 197 |
| GO:0003723 | RNA binding | 1.64e-01 | 1.00e+00 | 2.498 | 1 | 22 | 247 |
| GO:0003682 | chromatin binding | 1.67e-01 | 1.00e+00 | 2.469 | 1 | 15 | 252 |
| GO:0005634 | nucleus | 1.72e-01 | 1.00e+00 | 0.782 | 4 | 158 | 3246 |
| GO:0006351 | transcription, DNA-templated | 1.75e-01 | 1.00e+00 | 1.375 | 2 | 47 | 1076 |
| GO:0000278 | mitotic cell cycle | 2.05e-01 | 1.00e+00 | 2.151 | 1 | 28 | 314 |
| GO:0055085 | transmembrane transport | 2.13e-01 | 1.00e+00 | 2.084 | 1 | 13 | 329 |
| GO:0006366 | transcription from RNA polymerase II promoter | 2.20e-01 | 1.00e+00 | 2.032 | 1 | 23 | 341 |
| GO:0042803 | protein homodimerization activity | 2.69e-01 | 1.00e+00 | 1.705 | 1 | 13 | 428 |
| GO:0005737 | cytoplasm | 2.85e-01 | 1.00e+00 | 0.669 | 3 | 127 | 2633 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.85e-01 | 1.00e+00 | 1.613 | 1 | 29 | 456 |
| GO:0010467 | gene expression | 3.26e-01 | 1.00e+00 | 1.383 | 1 | 49 | 535 |
| GO:0005886 | plasma membrane | 3.75e-01 | 1.00e+00 | 0.645 | 2 | 46 | 1784 |
| GO:0008270 | zinc ion binding | 4.18e-01 | 1.00e+00 | 0.940 | 1 | 39 | 727 |
| GO:0044281 | small molecule metabolic process | 4.69e-01 | 1.00e+00 | 0.725 | 1 | 35 | 844 |
| GO:0046872 | metal ion binding | 5.05e-01 | 1.00e+00 | 0.579 | 1 | 24 | 934 |
| GO:0003677 | DNA binding | 5.10e-01 | 1.00e+00 | 0.559 | 1 | 52 | 947 |
| GO:0016020 | membrane | 6.03e-01 | 1.00e+00 | 0.209 | 1 | 56 | 1207 |
| GO:0005730 | nucleolus | 6.06e-01 | 1.00e+00 | 0.197 | 1 | 74 | 1217 |
| GO:0016021 | integral component of membrane | 6.96e-01 | 1.00e+00 | -0.129 | 1 | 19 | 1526 |
| GO:0005829 | cytosol | 7.57e-01 | 1.00e+00 | -0.357 | 1 | 88 | 1787 |