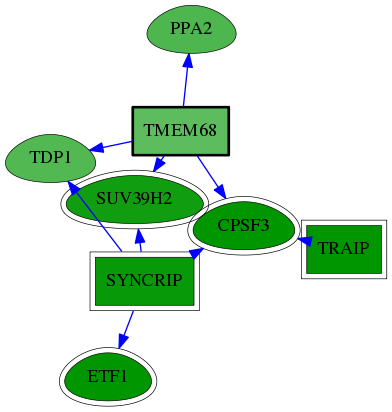
| GO:0003723 | RNA binding | 8.15e-04 | 1.00e+00 | 3.890 | 3 | 22 | 247 |
| GO:0017005 | 3'-tyrosyl-DNA phosphodiesterase activity | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0016149 | translation release factor activity, codon specific | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0008079 | translation termination factor activity | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0071344 | diphosphate metabolic process | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0004427 | inorganic diphosphatase activity | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0003747 | translation release factor activity | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 2 | 2 |
| GO:0030529 | ribonucleoprotein complex | 1.89e-03 | 1.00e+00 | 4.896 | 2 | 5 | 82 |
| GO:0036123 | histone H3-K9 dimethylation | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:0006449 | regulation of translational termination | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0000012 | single strand break repair | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0006333 | chromatin assembly or disassembly | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0036124 | histone H3-K9 trimethylation | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 2 | 4 |
| GO:0097452 | GAIT complex | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 2 | 4 |
| GO:0004521 | endoribonuclease activity | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0006398 | histone mRNA 3'-end processing | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 2 | 5 |
| GO:0000398 | mRNA splicing, via spliceosome | 4.53e-03 | 1.00e+00 | 4.253 | 2 | 15 | 128 |
| GO:0070937 | CRD-mediated mRNA stability complex | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 2 | 6 |
| GO:0042754 | negative regulation of circadian rhythm | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 2 | 6 |
| GO:0046974 | histone methyltransferase activity (H3-K9 specific) | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0007140 | male meiosis | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 7 |
| GO:0008409 | 5'-3' exonuclease activity | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 2 | 7 |
| GO:0006379 | mRNA cleavage | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 2 | 9 |
| GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 3 | 9 |
| GO:0008380 | RNA splicing | 7.43e-03 | 1.00e+00 | 3.887 | 2 | 21 | 165 |
| GO:0010467 | gene expression | 7.44e-03 | 1.00e+00 | 2.775 | 3 | 49 | 535 |
| GO:0008143 | poly(A) binding | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 2 | 10 |
| GO:0006479 | protein methylation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 11 |
| GO:0004527 | exonuclease activity | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 4 | 12 |
| GO:0016746 | transferase activity, transferring acyl groups | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 2 | 14 |
| GO:0043022 | ribosome binding | 1.38e-02 | 1.00e+00 | 6.166 | 1 | 2 | 17 |
| GO:0006378 | mRNA polyadenylation | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 3 | 18 |
| GO:0071346 | cellular response to interferon-gamma | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 2 | 20 |
| GO:0005720 | nuclear heterochromatin | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 3 | 20 |
| GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1.71e-02 | 1.00e+00 | 5.861 | 1 | 2 | 21 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 3 | 25 |
| GO:0005057 | receptor signaling protein activity | 2.11e-02 | 1.00e+00 | 5.553 | 1 | 2 | 26 |
| GO:0004722 | protein serine/threonine phosphatase activity | 2.27e-02 | 1.00e+00 | 5.446 | 1 | 1 | 28 |
| GO:0006418 | tRNA aminoacylation for protein translation | 2.35e-02 | 1.00e+00 | 5.395 | 1 | 4 | 29 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 2.51e-02 | 1.00e+00 | 5.299 | 1 | 6 | 31 |
| GO:0017148 | negative regulation of translation | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 5 | 33 |
| GO:0031124 | mRNA 3'-end processing | 2.83e-02 | 1.00e+00 | 5.124 | 1 | 4 | 35 |
| GO:0006369 | termination of RNA polymerase II transcription | 3.07e-02 | 1.00e+00 | 5.005 | 1 | 5 | 38 |
| GO:0048511 | rhythmic process | 3.07e-02 | 1.00e+00 | 5.005 | 1 | 5 | 38 |
| GO:0006396 | RNA processing | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 4 | 40 |
| GO:0000775 | chromosome, centromeric region | 3.31e-02 | 1.00e+00 | 4.896 | 1 | 6 | 41 |
| GO:0006406 | mRNA export from nucleus | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 7 | 44 |
| GO:0006302 | double-strand break repair | 4.18e-02 | 1.00e+00 | 4.553 | 1 | 5 | 52 |
| GO:0006338 | chromatin remodeling | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 7 | 55 |
| GO:0016311 | dephosphorylation | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 3 | 55 |
| GO:0003697 | single-stranded DNA binding | 4.58e-02 | 1.00e+00 | 4.421 | 1 | 4 | 57 |
| GO:0000785 | chromatin | 4.73e-02 | 1.00e+00 | 4.371 | 1 | 6 | 59 |
| GO:0071013 | catalytic step 2 spliceosome | 4.89e-02 | 1.00e+00 | 4.323 | 1 | 5 | 61 |
| GO:0006415 | translational termination | 5.05e-02 | 1.00e+00 | 4.276 | 1 | 2 | 63 |
| GO:0001649 | osteoblast differentiation | 5.91e-02 | 1.00e+00 | 4.044 | 1 | 5 | 74 |
| GO:0071456 | cellular response to hypoxia | 6.06e-02 | 1.00e+00 | 4.005 | 1 | 2 | 76 |
| GO:0005515 | protein binding | 6.50e-02 | 1.00e+00 | 0.829 | 6 | 198 | 4124 |
| GO:0003690 | double-stranded DNA binding | 6.60e-02 | 1.00e+00 | 3.878 | 1 | 6 | 83 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6.68e-02 | 1.00e+00 | 3.861 | 1 | 5 | 84 |
| GO:0000287 | magnesium ion binding | 9.05e-02 | 1.00e+00 | 3.408 | 1 | 4 | 115 |
| GO:0005759 | mitochondrial matrix | 1.08e-01 | 1.00e+00 | 3.134 | 1 | 7 | 139 |
| GO:0016874 | ligase activity | 1.14e-01 | 1.00e+00 | 3.064 | 1 | 11 | 146 |
| GO:0008270 | zinc ion binding | 1.15e-01 | 1.00e+00 | 1.748 | 2 | 39 | 727 |
| GO:0016071 | mRNA metabolic process | 1.31e-01 | 1.00e+00 | 2.844 | 1 | 10 | 170 |
| GO:0000166 | nucleotide binding | 1.35e-01 | 1.00e+00 | 2.794 | 1 | 13 | 176 |
| GO:0044822 | poly(A) RNA binding | 1.35e-01 | 1.00e+00 | 1.611 | 2 | 45 | 799 |
| GO:0006412 | translation | 1.37e-01 | 1.00e+00 | 2.778 | 1 | 12 | 178 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.38e-01 | 1.00e+00 | 2.770 | 1 | 17 | 179 |
| GO:0008152 | metabolic process | 1.40e-01 | 1.00e+00 | 2.746 | 1 | 6 | 182 |
| GO:0016070 | RNA metabolic process | 1.44e-01 | 1.00e+00 | 2.699 | 1 | 10 | 188 |
| GO:0016567 | protein ubiquitination | 1.44e-01 | 1.00e+00 | 2.699 | 1 | 14 | 188 |
| GO:0035556 | intracellular signal transduction | 1.52e-01 | 1.00e+00 | 2.617 | 1 | 5 | 199 |
| GO:0030154 | cell differentiation | 1.55e-01 | 1.00e+00 | 2.581 | 1 | 5 | 204 |
| GO:0006281 | DNA repair | 1.55e-01 | 1.00e+00 | 2.588 | 1 | 24 | 203 |
| GO:0005654 | nucleoplasm | 1.57e-01 | 1.00e+00 | 1.479 | 2 | 68 | 876 |
| GO:0008283 | cell proliferation | 1.80e-01 | 1.00e+00 | 2.353 | 1 | 8 | 239 |
| GO:0003682 | chromatin binding | 1.89e-01 | 1.00e+00 | 2.276 | 1 | 15 | 252 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.32e-01 | 1.00e+00 | 1.945 | 1 | 17 | 317 |
| GO:0006366 | transcription from RNA polymerase II promoter | 2.48e-01 | 1.00e+00 | 1.840 | 1 | 23 | 341 |
| GO:0048471 | perinuclear region of cytoplasm | 2.49e-01 | 1.00e+00 | 1.827 | 1 | 11 | 344 |
| GO:0044267 | cellular protein metabolic process | 2.55e-01 | 1.00e+00 | 1.794 | 1 | 14 | 352 |
| GO:0005783 | endoplasmic reticulum | 2.73e-01 | 1.00e+00 | 1.680 | 1 | 12 | 381 |
| GO:0006915 | apoptotic process | 2.91e-01 | 1.00e+00 | 1.570 | 1 | 15 | 411 |
| GO:0016032 | viral process | 3.01e-01 | 1.00e+00 | 1.512 | 1 | 30 | 428 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.18e-01 | 1.00e+00 | 1.421 | 1 | 29 | 456 |
| GO:0007165 | signal transduction | 4.07e-01 | 1.00e+00 | 0.982 | 1 | 24 | 618 |
| GO:0046872 | metal ion binding | 5.53e-01 | 1.00e+00 | 0.386 | 1 | 24 | 934 |
| GO:0006351 | transcription, DNA-templated | 6.07e-01 | 1.00e+00 | 0.182 | 1 | 47 | 1076 |
| GO:0016020 | membrane | 6.52e-01 | 1.00e+00 | 0.016 | 1 | 56 | 1207 |
| GO:0005730 | nucleolus | 6.55e-01 | 1.00e+00 | 0.004 | 1 | 74 | 1217 |
| GO:0005737 | cytoplasm | 6.80e-01 | 1.00e+00 | -0.109 | 2 | 127 | 2633 |
| GO:0016021 | integral component of membrane | 7.43e-01 | 1.00e+00 | -0.322 | 1 | 19 | 1526 |
| GO:0070062 | extracellular vesicular exosome | 7.71e-01 | 1.00e+00 | -0.427 | 1 | 51 | 1641 |
| GO:0005829 | cytosol | 8.02e-01 | 1.00e+00 | -0.550 | 1 | 88 | 1787 |
| GO:0005634 | nucleus | 8.04e-01 | 1.00e+00 | -0.411 | 2 | 158 | 3246 |