int-snw-3162
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 2.821 | 2.37e-15 | 5.75e-04 | 3.83e-03 |
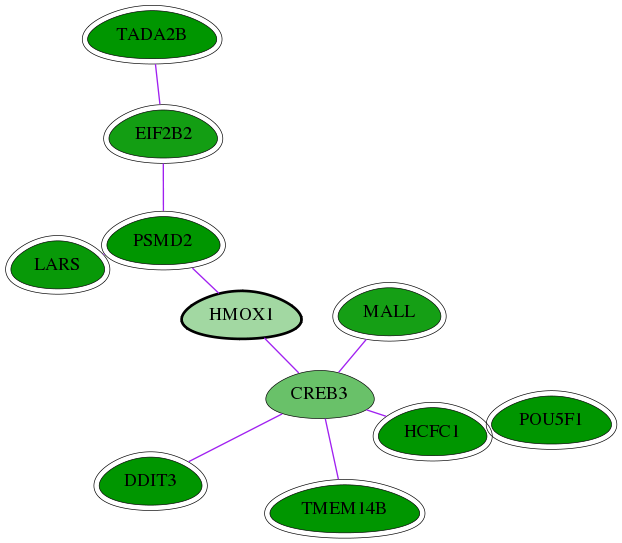
Genes (11)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| HCFC1 | 3054 | 40 | -2.925 | 3.449 | 71 | Yes | Yes |
| MALL | 7851 | 1 | -2.444 | 2.821 | 3 | Yes | - |
| LARS | 51520 | 14 | -2.574 | 3.157 | 105 | Yes | Yes |
| PSMD2 | 5708 | 47 | -4.172 | 3.157 | 386 | Yes | Yes |
| TADA2B | 93624 | 11 | -3.061 | 3.082 | 19 | Yes | - |
| EIF2B2 | 8892 | 8 | -2.462 | 2.968 | 95 | Yes | Yes |
| CREB3 | 10488 | 1 | -1.562 | 2.821 | 124 | - | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| [ HMOX1 ] | 3162 | 1 | -0.975 | 2.821 | 11 | - | - |
| DDIT3 | 1649 | 60 | -2.825 | 3.504 | 343 | Yes | Yes |
| TMEM14B | 81853 | 1 | -2.879 | 2.821 | 5 | Yes | Yes |
Interactions (10)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| HMOX1 | 3162 | CREB3 | 10488 | pp | -- | int.I2D: MINT |
| EIF2B2 | 8892 | TADA2B | 93624 | pp | -- | int.I2D: BioGrid_Fly, BIND_Fly, FlyLow, IntAct_Fly, MINT_Fly |
| HCFC1 | 3054 | CREB3 | 10488 | pp | -- | int.Intact: MI:0915(physical association), MI:0407(direct interaction); int.I2D: BioGrid, HPRD, IntAct, BCI; int.Ravasi: -; int.HPRD: in vitro, in vivo |
| PSMD2 | 5708 | LARS | 51520 | pp | -- | int.I2D: BioGrid_Yeast |
| HCFC1 | 3054 | POU5F1 | 5460 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| HMOX1 | 3162 | PSMD2 | 5708 | pp | -- | int.I2D: BioGrid |
| MALL | 7851 | CREB3 | 10488 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: HPRD, IntAct, BioGrid, MINT, VidalHuman_core; int.Mint: MI:0915(physical association); int.HPRD: yeast 2-hybrid |
| PSMD2 | 5708 | EIF2B2 | 8892 | pp | -- | int.I2D: IntAct_Yeast |
| CREB3 | 10488 | TMEM14B | 81853 | pp | -- | int.I2D: MINT |
| DDIT3 | 1649 | CREB3 | 10488 | pp | -- | int.I2D: BIND |
Related GO terms (232)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0019046 | release from viral latency | 5.29e-07 | 7.63e-03 | 10.357 | 2 | 2 | 2 |
| GO:0070461 | SAGA-type complex | 5.28e-06 | 7.62e-02 | 9.035 | 2 | 2 | 5 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 4.11e-05 | 5.92e-01 | 7.656 | 2 | 2 | 13 |
| GO:0006986 | response to unfolded protein | 6.35e-04 | 1.00e+00 | 5.713 | 2 | 4 | 50 |
| GO:0034976 | response to endoplasmic reticulum stress | 6.61e-04 | 1.00e+00 | 5.684 | 2 | 2 | 51 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:2000016 | negative regulation of determination of dorsal identity | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0043305 | negative regulation of mast cell degranulation | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:2000326 | negative regulation of ligand-dependent nuclear receptor transcription coactivator activity | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 7.63e-04 | 1.00e+00 | 10.357 | 1 | 1 | 1 |
| GO:0005829 | cytosol | 7.94e-04 | 1.00e+00 | 1.879 | 7 | 86 | 2496 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 8.84e-04 | 1.00e+00 | 5.474 | 2 | 6 | 59 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 1.37e-03 | 1.00e+00 | 2.908 | 4 | 39 | 699 |
| GO:0019043 | establishment of viral latency | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0006788 | heme oxidation | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 2 | 2 |
| GO:0006429 | leucyl-tRNA aminoacylation | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0032764 | negative regulation of mast cell cytokine production | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0002246 | wound healing involved in inflammatory response | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0014806 | smooth muscle hyperplasia | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0009786 | regulation of asymmetric cell division | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0004823 | leucine-tRNA ligase activity | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0004392 | heme oxygenase (decyclizing) activity | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 2 | 2 |
| GO:0044324 | regulation of transcription involved in anterior/posterior axis specification | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1.52e-03 | 1.00e+00 | 9.357 | 1 | 1 | 2 |
| GO:0003682 | chromatin binding | 1.66e-03 | 1.00e+00 | 3.588 | 3 | 19 | 327 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 2.15e-03 | 1.00e+00 | 2.733 | 4 | 41 | 789 |
| GO:0090045 | positive regulation of deacetylase activity | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 2 | 3 |
| GO:0031670 | cellular response to nutrient | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0060913 | cardiac cell fate determination | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron | 2.29e-03 | 1.00e+00 | 8.772 | 1 | 1 | 3 |
| GO:0051716 | cellular response to stimulus | 3.05e-03 | 1.00e+00 | 8.357 | 1 | 4 | 4 |
| GO:0046523 | S-methyl-5-thioribose-1-phosphate isomerase activity | 3.05e-03 | 1.00e+00 | 8.357 | 1 | 3 | 4 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.38e-03 | 1.00e+00 | 3.231 | 3 | 30 | 419 |
| GO:0006325 | chromatin organization | 3.48e-03 | 1.00e+00 | 4.474 | 2 | 12 | 118 |
| GO:0001714 | endodermal cell fate specification | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 1 | 5 |
| GO:0043620 | regulation of DNA-templated transcription in response to stress | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 1 | 5 |
| GO:0004630 | phospholipase D activity | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 1 | 5 |
| GO:0002686 | negative regulation of leukocyte migration | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 1 | 5 |
| GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 3.81e-03 | 1.00e+00 | 8.035 | 1 | 1 | 5 |
| GO:0042167 | heme catabolic process | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 2 | 6 |
| GO:0031726 | CCR1 chemokine receptor binding | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 6 |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 4 | 6 |
| GO:0010656 | negative regulation of muscle cell apoptotic process | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 6 |
| GO:0006355 | regulation of transcription, DNA-templated | 5.05e-03 | 1.00e+00 | 2.395 | 4 | 43 | 997 |
| GO:0071243 | cellular response to arsenic-containing substance | 5.33e-03 | 1.00e+00 | 7.549 | 1 | 1 | 7 |
| GO:0034383 | low-density lipoprotein particle clearance | 5.33e-03 | 1.00e+00 | 7.549 | 1 | 1 | 7 |
| GO:0034101 | erythrocyte homeostasis | 5.33e-03 | 1.00e+00 | 7.549 | 1 | 1 | 7 |
| GO:0002161 | aminoacyl-tRNA editing activity | 5.33e-03 | 1.00e+00 | 7.549 | 1 | 2 | 7 |
| GO:0001955 | blood vessel maturation | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 1 | 8 |
| GO:0008140 | cAMP response element binding protein binding | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 1 | 8 |
| GO:0070688 | MLL5-L complex | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 1 | 8 |
| GO:0043254 | regulation of protein complex assembly | 6.09e-03 | 1.00e+00 | 7.357 | 1 | 1 | 8 |
| GO:0005654 | nucleoplasm | 6.78e-03 | 1.00e+00 | 2.277 | 4 | 64 | 1082 |
| GO:0043995 | histone acetyltransferase activity (H4-K5 specific) | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 5 | 9 |
| GO:0046972 | histone acetyltransferase activity (H4-K16 specific) | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 5 | 9 |
| GO:0019509 | L-methionine biosynthetic process from methylthioadenosine | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 3 | 9 |
| GO:0048188 | Set1C/COMPASS complex | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 4 | 9 |
| GO:0006983 | ER overload response | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 1 | 9 |
| GO:0006450 | regulation of translational fidelity | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 2 | 9 |
| GO:0045080 | positive regulation of chemokine biosynthetic process | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 1 | 9 |
| GO:0043996 | histone acetyltransferase activity (H4-K8 specific) | 6.85e-03 | 1.00e+00 | 7.187 | 1 | 5 | 9 |
| GO:0044212 | transcription regulatory region DNA binding | 6.92e-03 | 1.00e+00 | 3.964 | 2 | 17 | 168 |
| GO:0035413 | positive regulation of catenin import into nucleus | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 2 | 10 |
| GO:0000982 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 1 | 10 |
| GO:0090026 | positive regulation of monocyte chemotaxis | 8.36e-03 | 1.00e+00 | 6.897 | 1 | 1 | 11 |
| GO:0045662 | negative regulation of myoblast differentiation | 8.36e-03 | 1.00e+00 | 6.897 | 1 | 3 | 11 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 8.36e-03 | 1.00e+00 | 6.897 | 1 | 4 | 11 |
| GO:0005838 | proteasome regulatory particle | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 12 |
| GO:0001935 | endothelial cell proliferation | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 12 |
| GO:0035198 | miRNA binding | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 12 |
| GO:0030914 | STAGA complex | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 3 | 13 |
| GO:0042994 | cytoplasmic sequestering of transcription factor | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 13 |
| GO:0030234 | enzyme regulator activity | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 2 | 13 |
| GO:0071276 | cellular response to cadmium ion | 9.87e-03 | 1.00e+00 | 6.656 | 1 | 1 | 13 |
| GO:0003677 | DNA binding | 1.03e-02 | 1.00e+00 | 2.106 | 4 | 49 | 1218 |
| GO:0050930 | induction of positive chemotaxis | 1.06e-02 | 1.00e+00 | 6.549 | 1 | 1 | 14 |
| GO:0043981 | histone H4-K5 acetylation | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 5 | 15 |
| GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 3 | 15 |
| GO:0001824 | blastocyst development | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 2 | 15 |
| GO:0043982 | histone H4-K8 acetylation | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 5 | 15 |
| GO:0002230 | positive regulation of defense response to virus by host | 1.14e-02 | 1.00e+00 | 6.450 | 1 | 1 | 15 |
| GO:0042176 | regulation of protein catabolic process | 1.21e-02 | 1.00e+00 | 6.357 | 1 | 2 | 16 |
| GO:0010467 | gene expression | 1.24e-02 | 1.00e+00 | 2.556 | 3 | 45 | 669 |
| GO:0022624 | proteasome accessory complex | 1.29e-02 | 1.00e+00 | 6.269 | 1 | 1 | 17 |
| GO:0006778 | porphyrin-containing compound metabolic process | 1.36e-02 | 1.00e+00 | 6.187 | 1 | 2 | 18 |
| GO:0008134 | transcription factor binding | 1.40e-02 | 1.00e+00 | 3.438 | 2 | 18 | 242 |
| GO:0043025 | neuronal cell body | 1.43e-02 | 1.00e+00 | 3.420 | 2 | 9 | 245 |
| GO:0043984 | histone H4-K16 acetylation | 1.44e-02 | 1.00e+00 | 6.109 | 1 | 5 | 19 |
| GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 3 | 20 |
| GO:0014003 | oligodendrocyte development | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 5 | 20 |
| GO:0032757 | positive regulation of interleukin-8 production | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 1 | 20 |
| GO:0055072 | iron ion homeostasis | 1.59e-02 | 1.00e+00 | 5.964 | 1 | 2 | 21 |
| GO:0000123 | histone acetyltransferase complex | 1.59e-02 | 1.00e+00 | 5.964 | 1 | 6 | 21 |
| GO:0045787 | positive regulation of cell cycle | 1.67e-02 | 1.00e+00 | 5.897 | 1 | 2 | 22 |
| GO:0001205 | RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 1.67e-02 | 1.00e+00 | 5.897 | 1 | 4 | 22 |
| GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1.74e-02 | 1.00e+00 | 5.833 | 1 | 1 | 23 |
| GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1.74e-02 | 1.00e+00 | 5.833 | 1 | 1 | 23 |
| GO:0045765 | regulation of angiogenesis | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 2 | 24 |
| GO:0035094 | response to nicotine | 1.89e-02 | 1.00e+00 | 5.713 | 1 | 2 | 25 |
| GO:0043392 | negative regulation of DNA binding | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 4 | 26 |
| GO:0045909 | positive regulation of vasodilation | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 1 | 26 |
| GO:0051209 | release of sequestered calcium ion into cytosol | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 1 | 26 |
| GO:0051928 | positive regulation of calcium ion transport | 1.97e-02 | 1.00e+00 | 5.656 | 1 | 1 | 26 |
| GO:0071339 | MLL1 complex | 2.04e-02 | 1.00e+00 | 5.602 | 1 | 6 | 27 |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | 2.12e-02 | 1.00e+00 | 5.549 | 1 | 2 | 28 |
| GO:0008219 | cell death | 2.12e-02 | 1.00e+00 | 5.549 | 1 | 2 | 28 |
| GO:0016604 | nuclear body | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 3 | 31 |
| GO:0042594 | response to starvation | 2.34e-02 | 1.00e+00 | 5.402 | 1 | 4 | 31 |
| GO:0042552 | myelination | 2.64e-02 | 1.00e+00 | 5.227 | 1 | 4 | 35 |
| GO:0007588 | excretion | 2.64e-02 | 1.00e+00 | 5.227 | 1 | 2 | 35 |
| GO:0006446 | regulation of translational initiation | 2.71e-02 | 1.00e+00 | 5.187 | 1 | 4 | 36 |
| GO:0043565 | sequence-specific DNA binding | 2.74e-02 | 1.00e+00 | 2.922 | 2 | 11 | 346 |
| GO:0001541 | ovarian follicle development | 2.79e-02 | 1.00e+00 | 5.147 | 1 | 2 | 37 |
| GO:0042542 | response to hydrogen peroxide | 2.94e-02 | 1.00e+00 | 5.071 | 1 | 1 | 39 |
| GO:0045786 | negative regulation of cell cycle | 2.94e-02 | 1.00e+00 | 5.071 | 1 | 3 | 39 |
| GO:0000139 | Golgi membrane | 2.96e-02 | 1.00e+00 | 2.861 | 2 | 7 | 361 |
| GO:0009408 | response to heat | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 6 | 40 |
| GO:0035019 | somatic stem cell maintenance | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 6 | 40 |
| GO:0016020 | membrane | 3.09e-02 | 1.00e+00 | 1.642 | 4 | 46 | 1681 |
| GO:0030136 | clathrin-coated vesicle | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 5 | 42 |
| GO:0006418 | tRNA aminoacylation for protein translation | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 3 | 42 |
| GO:0043434 | response to peptide hormone | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 5 | 45 |
| GO:0043525 | positive regulation of neuron apoptotic process | 3.45e-02 | 1.00e+00 | 4.833 | 1 | 4 | 46 |
| GO:0042632 | cholesterol homeostasis | 3.60e-02 | 1.00e+00 | 4.772 | 1 | 1 | 48 |
| GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 3.68e-02 | 1.00e+00 | 4.742 | 1 | 2 | 49 |
| GO:0048661 | positive regulation of smooth muscle cell proliferation | 3.68e-02 | 1.00e+00 | 4.742 | 1 | 1 | 49 |
| GO:0003743 | translation initiation factor activity | 3.68e-02 | 1.00e+00 | 4.742 | 1 | 7 | 49 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 3.75e-02 | 1.00e+00 | 4.713 | 1 | 1 | 50 |
| GO:0045454 | cell redox homeostasis | 3.82e-02 | 1.00e+00 | 4.684 | 1 | 1 | 51 |
| GO:0006879 | cellular iron ion homeostasis | 3.82e-02 | 1.00e+00 | 4.684 | 1 | 3 | 51 |
| GO:0005515 | protein binding | 3.84e-02 | 1.00e+00 | 0.800 | 8 | 198 | 6024 |
| GO:0009611 | response to wounding | 3.90e-02 | 1.00e+00 | 4.656 | 1 | 5 | 52 |
| GO:0030176 | integral component of endoplasmic reticulum membrane | 3.97e-02 | 1.00e+00 | 4.629 | 1 | 1 | 53 |
| GO:0043627 | response to estrogen | 4.19e-02 | 1.00e+00 | 4.549 | 1 | 5 | 56 |
| GO:0008217 | regulation of blood pressure | 4.26e-02 | 1.00e+00 | 4.524 | 1 | 1 | 57 |
| GO:0000502 | proteasome complex | 4.34e-02 | 1.00e+00 | 4.499 | 1 | 3 | 58 |
| GO:0001558 | regulation of cell growth | 4.56e-02 | 1.00e+00 | 4.426 | 1 | 1 | 61 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 4.56e-02 | 1.00e+00 | 4.426 | 1 | 2 | 61 |
| GO:0005901 | caveola | 4.63e-02 | 1.00e+00 | 4.402 | 1 | 1 | 62 |
| GO:0009749 | response to glucose | 4.78e-02 | 1.00e+00 | 4.357 | 1 | 7 | 64 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 4.78e-02 | 1.00e+00 | 4.357 | 1 | 1 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4.85e-02 | 1.00e+00 | 4.334 | 1 | 2 | 65 |
| GO:0044267 | cellular protein metabolic process | 4.87e-02 | 1.00e+00 | 2.468 | 2 | 20 | 474 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 4.95e-02 | 1.00e+00 | 2.456 | 2 | 24 | 478 |
| GO:0010468 | regulation of gene expression | 5.07e-02 | 1.00e+00 | 4.269 | 1 | 2 | 68 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.21e-02 | 1.00e+00 | 4.227 | 1 | 2 | 70 |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 5.29e-02 | 1.00e+00 | 4.207 | 1 | 5 | 71 |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 5.29e-02 | 1.00e+00 | 4.207 | 1 | 4 | 71 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 5.36e-02 | 1.00e+00 | 4.187 | 1 | 1 | 72 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 5.50e-02 | 1.00e+00 | 4.147 | 1 | 4 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.50e-02 | 1.00e+00 | 4.147 | 1 | 2 | 74 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 5.58e-02 | 1.00e+00 | 4.128 | 1 | 1 | 75 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 5.58e-02 | 1.00e+00 | 4.128 | 1 | 1 | 75 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 5.86e-02 | 1.00e+00 | 4.053 | 1 | 2 | 79 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 5.86e-02 | 1.00e+00 | 4.053 | 1 | 2 | 79 |
| GO:0016032 | viral process | 6.03e-02 | 1.00e+00 | 2.296 | 2 | 26 | 534 |
| GO:0009653 | anatomical structure morphogenesis | 6.15e-02 | 1.00e+00 | 3.982 | 1 | 2 | 83 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 6.37e-02 | 1.00e+00 | 3.930 | 1 | 8 | 86 |
| GO:0006979 | response to oxidative stress | 6.44e-02 | 1.00e+00 | 3.914 | 1 | 6 | 87 |
| GO:0005783 | endoplasmic reticulum | 6.48e-02 | 1.00e+00 | 2.238 | 2 | 13 | 556 |
| GO:0050821 | protein stabilization | 6.51e-02 | 1.00e+00 | 3.897 | 1 | 3 | 88 |
| GO:0045766 | positive regulation of angiogenesis | 6.51e-02 | 1.00e+00 | 3.897 | 1 | 2 | 88 |
| GO:0005789 | endoplasmic reticulum membrane | 6.77e-02 | 1.00e+00 | 2.202 | 2 | 9 | 570 |
| GO:0071456 | cellular response to hypoxia | 6.80e-02 | 1.00e+00 | 3.833 | 1 | 3 | 92 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 6.80e-02 | 1.00e+00 | 3.833 | 1 | 1 | 92 |
| GO:0005770 | late endosome | 6.87e-02 | 1.00e+00 | 3.817 | 1 | 2 | 93 |
| GO:0020037 | heme binding | 6.87e-02 | 1.00e+00 | 3.817 | 1 | 1 | 93 |
| GO:0007417 | central nervous system development | 7.09e-02 | 1.00e+00 | 3.772 | 1 | 5 | 96 |
| GO:0042803 | protein homodimerization activity | 7.30e-02 | 1.00e+00 | 2.140 | 2 | 19 | 595 |
| GO:0006935 | chemotaxis | 7.73e-02 | 1.00e+00 | 3.642 | 1 | 3 | 105 |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | 7.73e-02 | 1.00e+00 | 3.642 | 1 | 8 | 105 |
| GO:0042127 | regulation of cell proliferation | 8.01e-02 | 1.00e+00 | 3.588 | 1 | 4 | 109 |
| GO:0000209 | protein polyubiquitination | 8.50e-02 | 1.00e+00 | 3.499 | 1 | 3 | 116 |
| GO:0043524 | negative regulation of neuron apoptotic process | 8.57e-02 | 1.00e+00 | 3.486 | 1 | 5 | 117 |
| GO:0046983 | protein dimerization activity | 8.64e-02 | 1.00e+00 | 3.474 | 1 | 5 | 118 |
| GO:0007050 | cell cycle arrest | 9.06e-02 | 1.00e+00 | 3.402 | 1 | 5 | 124 |
| GO:0030335 | positive regulation of cell migration | 9.27e-02 | 1.00e+00 | 3.368 | 1 | 5 | 127 |
| GO:0006413 | translational initiation | 9.55e-02 | 1.00e+00 | 3.323 | 1 | 7 | 131 |
| GO:0005634 | nucleus | 9.78e-02 | 1.00e+00 | 0.787 | 6 | 159 | 4559 |
| GO:0016055 | Wnt signaling pathway | 1.00e-01 | 1.00e+00 | 3.248 | 1 | 4 | 138 |
| GO:0051260 | protein homooligomerization | 1.05e-01 | 1.00e+00 | 3.177 | 1 | 3 | 145 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 1.05e-01 | 1.00e+00 | 3.177 | 1 | 4 | 145 |
| GO:0010628 | positive regulation of gene expression | 1.06e-01 | 1.00e+00 | 3.167 | 1 | 7 | 146 |
| GO:0001077 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 1.07e-01 | 1.00e+00 | 3.157 | 1 | 6 | 147 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.08e-01 | 1.00e+00 | 3.137 | 1 | 3 | 149 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.09e-01 | 1.00e+00 | 3.118 | 1 | 7 | 151 |
| GO:0042981 | regulation of apoptotic process | 1.09e-01 | 1.00e+00 | 3.128 | 1 | 4 | 150 |
| GO:0045121 | membrane raft | 1.13e-01 | 1.00e+00 | 3.071 | 1 | 3 | 156 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.19e-01 | 1.00e+00 | 2.990 | 1 | 12 | 165 |
| GO:0034641 | cellular nitrogen compound metabolic process | 1.23e-01 | 1.00e+00 | 2.939 | 1 | 2 | 171 |
| GO:0007049 | cell cycle | 1.25e-01 | 1.00e+00 | 2.914 | 1 | 6 | 174 |
| GO:0005667 | transcription factor complex | 1.26e-01 | 1.00e+00 | 2.905 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 1.28e-01 | 1.00e+00 | 2.881 | 1 | 5 | 178 |
| GO:0003714 | transcription corepressor activity | 1.28e-01 | 1.00e+00 | 2.881 | 1 | 11 | 178 |
| GO:0043547 | positive regulation of GTPase activity | 1.29e-01 | 1.00e+00 | 2.865 | 1 | 9 | 180 |
| GO:0005737 | cytoplasm | 1.33e-01 | 1.00e+00 | 0.799 | 5 | 124 | 3767 |
| GO:0001525 | angiogenesis | 1.36e-01 | 1.00e+00 | 2.779 | 1 | 7 | 191 |
| GO:0004871 | signal transducer activity | 1.48e-01 | 1.00e+00 | 2.656 | 1 | 4 | 208 |
| GO:0016071 | mRNA metabolic process | 1.58e-01 | 1.00e+00 | 2.556 | 1 | 8 | 223 |
| GO:0006412 | translation | 1.62e-01 | 1.00e+00 | 2.511 | 1 | 8 | 230 |
| GO:0003713 | transcription coactivator activity | 1.67e-01 | 1.00e+00 | 2.468 | 1 | 24 | 237 |
| GO:0016070 | RNA metabolic process | 1.73e-01 | 1.00e+00 | 2.408 | 1 | 8 | 247 |
| GO:0016021 | integral component of membrane | 1.84e-01 | 1.00e+00 | 0.989 | 3 | 17 | 1982 |
| GO:0019899 | enzyme binding | 1.92e-01 | 1.00e+00 | 2.243 | 1 | 9 | 277 |
| GO:0007264 | small GTPase mediated signal transduction | 1.93e-01 | 1.00e+00 | 2.233 | 1 | 3 | 279 |
| GO:0035556 | intracellular signal transduction | 2.08e-01 | 1.00e+00 | 2.113 | 1 | 9 | 303 |
| GO:0005525 | GTP binding | 2.13e-01 | 1.00e+00 | 2.081 | 1 | 12 | 310 |
| GO:0044281 | small molecule metabolic process | 2.35e-01 | 1.00e+00 | 1.115 | 2 | 20 | 1211 |
| GO:0046982 | protein heterodimerization activity | 2.55e-01 | 1.00e+00 | 1.787 | 1 | 8 | 380 |
| GO:0005524 | ATP binding | 2.60e-01 | 1.00e+00 | 1.015 | 2 | 31 | 1298 |
| GO:0000278 | mitotic cell cycle | 2.61e-01 | 1.00e+00 | 1.746 | 1 | 15 | 391 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.74e-01 | 1.00e+00 | 1.667 | 1 | 24 | 413 |
| GO:0043066 | negative regulation of apoptotic process | 2.80e-01 | 1.00e+00 | 1.629 | 1 | 16 | 424 |
| GO:0055085 | transmembrane transport | 2.86e-01 | 1.00e+00 | 1.592 | 1 | 8 | 435 |
| GO:0006351 | transcription, DNA-templated | 3.04e-01 | 1.00e+00 | 0.859 | 2 | 57 | 1446 |
| GO:0042802 | identical protein binding | 3.13e-01 | 1.00e+00 | 1.438 | 1 | 19 | 484 |
| GO:0048471 | perinuclear region of cytoplasm | 3.23e-01 | 1.00e+00 | 1.385 | 1 | 8 | 502 |
| GO:0006915 | apoptotic process | 3.51e-01 | 1.00e+00 | 1.240 | 1 | 12 | 555 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.58e-01 | 1.00e+00 | 1.202 | 1 | 37 | 570 |
| GO:0005730 | nucleolus | 3.61e-01 | 1.00e+00 | 0.676 | 2 | 66 | 1641 |
| GO:0005794 | Golgi apparatus | 3.78e-01 | 1.00e+00 | 1.104 | 1 | 9 | 610 |
| GO:0005615 | extracellular space | 5.30e-01 | 1.00e+00 | 0.454 | 1 | 20 | 957 |
| GO:0008270 | zinc ion binding | 5.45e-01 | 1.00e+00 | 0.395 | 1 | 27 | 997 |
| GO:0005739 | mitochondrion | 5.46e-01 | 1.00e+00 | 0.394 | 1 | 23 | 998 |
| GO:0044822 | poly(A) RNA binding | 5.67e-01 | 1.00e+00 | 0.312 | 1 | 50 | 1056 |
| GO:0046872 | metal ion binding | 6.48e-01 | 1.00e+00 | 0.005 | 1 | 29 | 1307 |
| GO:0070062 | extracellular vesicular exosome | 8.65e-01 | 1.00e+00 | -0.872 | 1 | 57 | 2400 |
| GO:0005886 | plasma membrane | 8.86e-01 | 1.00e+00 | -0.978 | 1 | 49 | 2582 |