meta-reg-snw-1432
Interacting subnets
| Subnetwork | Dataset | Score | p-value 1 | p-value 2 | p-value 3 | Size | Highlighted genes |
|---|---|---|---|---|---|---|---|
| int-snw-56848 | tai-screen-luciferase | 6.375 | 2.94e-136 | 3.59e-07 | 2.58e-03 | 24 | 19 |
| reg-snw-1432 | tai-screen-luciferase | 4.670 | 5.00e-24 | 6.11e-04 | 9.80e-04 | 9 | 5 |
| int-snw-6150 | tai-screen-luciferase | 6.375 | 3.15e-136 | 3.60e-07 | 2.59e-03 | 23 | 17 |
| int-snw-4831 | tai-screen-luciferase | 6.314 | 3.38e-133 | 5.36e-07 | 3.08e-03 | 24 | 20 |
| int-snw-51741 | tai-screen-luciferase | 6.174 | 2.67e-126 | 1.32e-06 | 4.54e-03 | 24 | 19 |
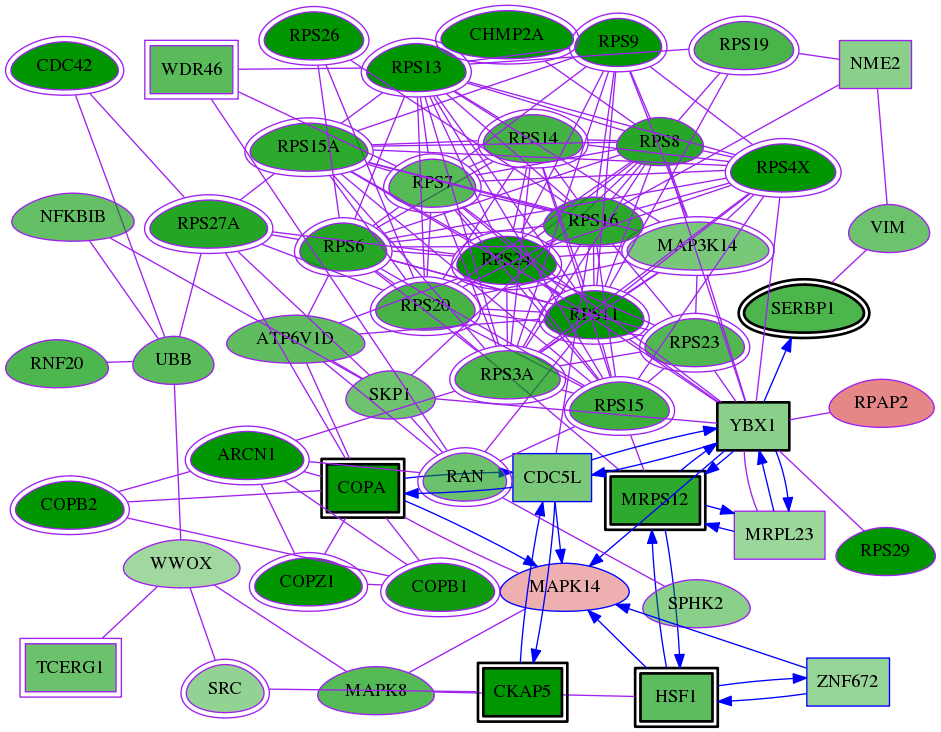
Genes (51)
| Gene Symbol | Entrez Gene ID | Frequency | tai-screen-luciferase gene score | Best subnetwork score | Degree | List-Gonzales GI | Tai-Hits |
|---|---|---|---|---|---|---|---|
| RPS20 | 6224 | 22 | -4.757 | 6.314 | 153 | Yes | - |
| RPS8 | 6202 | 33 | -5.545 | 7.555 | 234 | - | - |
| MAPK8 | 5599 | 28 | -4.403 | 6.468 | 153 | - | - |
| CHMP2A | 27243 | 33 | -9.037 | 7.555 | 41 | Yes | Yes |
| RPS15 | 6209 | 30 | -5.040 | 7.011 | 116 | Yes | - |
| COPA | 1314 | 48 | -9.395 | 5.672 | 340 | Yes | Yes |
| RNF20 | 56254 | 28 | -4.564 | 6.174 | 18 | - | - |
| RPS29 | 6235 | 17 | -8.386 | 6.597 | 29 | - | - |
| HSF1 | 3297 | 46 | -4.179 | 5.027 | 209 | - | Yes |
| MAPK14 | 1432 | 5 | 2.079 | 4.670 | 32 | - | - |
| SRC | 6714 | 28 | -2.806 | 6.174 | 419 | Yes | - |
| COPB2 | 9276 | 48 | -13.168 | 9.063 | 41 | Yes | Yes |
| CKAP5 | 9793 | 46 | -7.214 | 5.672 | 130 | Yes | Yes |
| RPS16 | 6217 | 38 | -5.444 | 4.880 | 205 | - | - |
| CDC42 | 998 | 44 | -6.960 | 4.707 | 276 | Yes | Yes |
| RPS11 | 6205 | 44 | -6.588 | 7.555 | 175 | Yes | - |
| NME2 | 4831 | 24 | -3.034 | 4.325 | 53 | - | - |
| RPS6 | 6194 | 44 | -5.603 | 8.046 | 217 | Yes | - |
| MRPS12 | 6183 | 35 | -5.421 | 5.516 | 341 | Yes | - |
| YBX1 | 4904 | 24 | -3.033 | 5.516 | 296 | - | - |
| RAN | 5901 | 38 | -3.809 | 4.325 | 258 | Yes | Yes |
| UBB | 7314 | 30 | -4.289 | 6.428 | 147 | - | - |
| MAP3K14 | 9020 | 15 | -3.462 | 6.375 | 138 | - | Yes |
| COPB1 | 1315 | 39 | -6.221 | 9.063 | 118 | Yes | Yes |
| SERBP1 | 26135 | 35 | -4.612 | 5.516 | 106 | - | Yes |
| RPS23 | 6228 | 21 | -4.485 | 6.375 | 118 | Yes | - |
| RPS13 | 6207 | 43 | -6.589 | 7.555 | 174 | Yes | - |
| CDC5L | 988 | 34 | -3.419 | 5.672 | 155 | - | - |
| ZNF672 | 79894 | 5 | -2.681 | 4.670 | 98 | - | - |
| RPS24 | 6229 | 46 | -7.034 | 8.389 | 217 | Yes | - |
| VIM | 7431 | 18 | -3.760 | 6.428 | 246 | - | - |
| RPAP2 | 79871 | 15 | 3.111 | 6.375 | 49 | - | - |
| TCERG1 | 10915 | 28 | -3.808 | 6.174 | 58 | Yes | - |
| RPS9 | 6203 | 45 | -7.127 | 7.555 | 140 | Yes | - |
| RPS3A | 6189 | 19 | -4.647 | 7.189 | 166 | Yes | - |
| RPS4X | 6191 | 44 | -6.747 | 7.555 | 263 | Yes | - |
| COPZ1 | 22818 | 48 | -8.301 | 9.063 | 13 | Yes | Yes |
| ATP6V1D | 51382 | 30 | -4.131 | 7.286 | 149 | - | - |
| RPS15A | 6210 | 36 | -5.413 | 7.555 | 177 | Yes | - |
| WDR46 | 9277 | 31 | -4.290 | 4.226 | 101 | Yes | Yes |
| WWOX | 51741 | 28 | -2.448 | 6.174 | 38 | - | - |
| RPS7 | 6201 | 15 | -4.382 | 6.375 | 165 | Yes | - |
| RPS27A | 6233 | 45 | -5.631 | 8.389 | 344 | Yes | - |
| RPS14 | 6208 | 23 | -4.803 | 7.011 | 204 | Yes | - |
| RPS19 | 6223 | 21 | -4.701 | 4.752 | 102 | Yes | - |
| SKP1 | 6500 | 26 | -3.750 | 6.413 | 203 | - | - |
| NFKBIB | 4793 | 28 | -3.978 | 6.174 | 78 | - | - |
| SPHK2 | 56848 | 17 | -3.010 | 6.375 | 37 | - | - |
| RPS26 | 6231 | 43 | -7.478 | 8.046 | 60 | Yes | - |
| ARCN1 | 372 | 48 | -8.232 | 9.063 | 118 | Yes | Yes |
| MRPL23 | 6150 | 15 | -2.571 | 6.375 | 9 | - | - |
Interactions (183)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| CDC42 | 998 | RPS27A | 6233 | pp | -- | int.I2D: SOURAV_MAPK_HIGH |
| RPS20 | 6224 | RPS26 | 6231 | pp | -- | int.I2D: YeastLow |
| RPS4X | 6191 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS27A | 6233 | SKP1 | 6500 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| YBX1 | 4904 | RPS23 | 6228 | pp | -- | int.Intact: MI:0914(association) |
| NME2 | 4831 | RPS19 | 6223 | pp | -- | int.Proteinpedia: Mass spectrometry |
| ARCN1 | 372 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, Tarassov_PCA, YeastMedium, INTEROLOG |
| RPS20 | 6224 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS13 | 6207 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BCI, HPRD, IntAct, MINT; int.Mint: MI:0915(physical association); int.HPRD: in vitro |
| VIM | 7431 | SERBP1 | 26135 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, HPRD, BioGrid, MINT, StelzlHigh; int.Mint: MI:0915(physical association); int.HPRD: yeast 2-hybrid |
| RPS11 | 6205 | RPS15 | 6209 | pp | -- | int.I2D: INTEROLOG, YeastMedium |
| RPS11 | 6205 | RPS19 | 6223 | pp | -- | int.I2D: IntAct_Yeast |
| RPS7 | 6201 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS11 | 6205 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| TCERG1 | 10915 | WWOX | 51741 | pp | -- | int.I2D: BioGrid, MINT, Pawson1; int.Mint: MI:0915(physical association) |
| RPS3A | 6189 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | RPS20 | 6224 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS11 | 6205 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS14 | 6208 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| SRC | 6714 | CKAP5 | 9793 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| YBX1 | 4904 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS6 | 6194 | RPS26 | 6231 | pp | -- | int.I2D: YeastLow |
| YBX1 | 4904 | RPS7 | 6201 | pp | -- | int.Intact: MI:0914(association) |
| ARCN1 | 372 | RAN | 5901 | pp | -- | int.I2D: YeastLow |
| RPS15 | 6209 | RPS23 | 6228 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastMedium |
| RPS7 | 6201 | RPS15 | 6209 | pp | -- | int.I2D: BioGrid_Yeast |
| HSF1 | 3297 | ZNF672 | 79894 | pd | <> | reg.ITFP.txt: no annot |
| YBX1 | 4904 | RPS11 | 6205 | pp | -- | int.Intact: MI:0914(association) |
| RPS14 | 6208 | RPS20 | 6224 | pp | -- | int.I2D: INTEROLOG, YeastMedium, BioGrid_Yeast |
| CDC42 | 998 | UBB | 7314 | pp | -- | int.I2D: SOURAV_MAPK_HIGH |
| NFKBIB | 4793 | UBB | 7314 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| RPS15 | 6209 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS9 | 6203 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS14 | 6208 | RPS15 | 6209 | pp | -- | int.I2D: INTEROLOG, YeastMedium |
| RPS8 | 6202 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS4X | 6191 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastMedium |
| YBX1 | 4904 | SERBP1 | 26135 | pd | > | reg.ITFP.txt: no annot |
| RPS9 | 6203 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS20 | 6224 | RPS23 | 6228 | pp | -- | int.I2D: YeastMedium, INTEROLOG |
| NME2 | 4831 | RPS16 | 6217 | pp | -- | int.Proteinpedia: Mass spectrometry |
| RPS8 | 6202 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS16 | 6217 | RPS20 | 6224 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS9 | 6203 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| MRPS12 | 6183 | RPS20 | 6224 | pp | -- | int.I2D: YeastMedium |
| SKP1 | 6500 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| RPS6 | 6194 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS13 | 6207 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS15 | 6209 | RPS20 | 6224 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastMedium |
| YBX1 | 4904 | RPS4X | 6191 | pp | -- | int.Intact: MI:0914(association) |
| RPS11 | 6205 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS23 | 6228 | pp | -- | int.I2D: YeastLow, BioGrid_Yeast |
| RPS24 | 6229 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS4X | 6191 | RPS7 | 6201 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS9 | 6203 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS15A | 6210 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| CDC5L | 988 | MAPK14 | 1432 | pd | > | reg.ITFP.txt: no annot |
| RAN | 5901 | RPS11 | 6205 | pp | -- | int.I2D: IntAct_Worm, BioGrid_Worm, BIND_Worm, MINT_Worm, NON_CORE |
| RPS7 | 6201 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | WDR46 | 9277 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS15 | 6209 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS14 | 6208 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS7 | 6201 | RPS9 | 6203 | pp | -- | int.I2D: BioGrid_Yeast |
| MAPK14 | 1432 | YBX1 | 4904 | pd | < | reg.ITFP.txt: no annot |
| RPS3A | 6189 | RPS23 | 6228 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS9 | 6203 | pp | -- | int.I2D: IntAct |
| RPS9 | 6203 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS14 | 6208 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| COPA | 1314 | MAPK14 | 1432 | pd | > | reg.ITFP.txt: no annot |
| COPA | 1314 | MAPK14 | 1432 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS4X | 6191 | RPS9 | 6203 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS8 | 6202 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| UBB | 7314 | WWOX | 51741 | pp | -- | int.I2D: BCI, HPRD; int.HPRD: in vivo |
| UBB | 7314 | RNF20 | 56254 | pp | -- | int.Intact: MI:0220(ubiquitination reaction); int.I2D: IntAct |
| MRPS12 | 6183 | RPS15 | 6209 | pp | -- | int.I2D: YeastMedium |
| RPS8 | 6202 | SKP1 | 6500 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS15A | 6210 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS11 | 6205 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS7 | 6201 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS11 | 6205 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| YBX1 | 4904 | SKP1 | 6500 | pp | -- | int.I2D: MINT; int.Mint: MI:0915(physical association) |
| RPS4X | 6191 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS8 | 6202 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| SRC | 6714 | WWOX | 51741 | pp | -- | int.I2D: HPRD; int.HPRD: in vitro, in vivo |
| RPS16 | 6217 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS7 | 6201 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS15A | 6210 | RPS20 | 6224 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastHigh, YeastMedium |
| MRPL23 | 6150 | MRPS12 | 6183 | pd | <> | reg.ITFP.txt: no annot |
| MAPK14 | 1432 | HSF1 | 3297 | pd | < | reg.ITFP.txt: no annot |
| YBX1 | 4904 | RPS8 | 6202 | pp | -- | int.Intact: MI:0914(association) |
| RPS13 | 6207 | RPS19 | 6223 | pp | -- | int.I2D: IntAct_Yeast |
| RPS20 | 6224 | RPS27A | 6233 | pp | -- | int.I2D: YeastLow |
| RPS4X | 6191 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS7 | 6201 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| MAPK14 | 1432 | MAPK8 | 5599 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| RPS27A | 6233 | UBB | 7314 | pp | -- | int.I2D: BIND |
| RPS4X | 6191 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| NME2 | 4831 | VIM | 7431 | pp | -- | int.HPRD: in vivo |
| RAN | 5901 | RPS15 | 6209 | pp | -- | int.I2D: YeastLow |
| RPS4X | 6191 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium, INTEROLOG |
| RPS8 | 6202 | RPS9 | 6203 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastMedium |
| RPS8 | 6202 | RPS19 | 6223 | pp | -- | int.I2D: MINT |
| RPS6 | 6194 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS27A | 6233 | pp | -- | int.I2D: YeastLow |
| RPS8 | 6202 | RPS20 | 6224 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS4X | 6191 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| COPA | 1314 | COPB1 | 1315 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct_Yeast, MINT_Yeast, Krogan_Core, YeastHigh |
| RPS8 | 6202 | CHMP2A | 27243 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS24 | 6229 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS7 | 6201 | RPS20 | 6224 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS7 | 6201 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| CDC5L | 988 | CKAP5 | 9793 | pd | <> | reg.ITFP.txt: no annot |
| RAN | 5901 | WDR46 | 9277 | pp | -- | int.I2D: YeastLow |
| YBX1 | 4904 | MRPL23 | 6150 | pd | <> | reg.ITFP.txt: no annot |
| YBX1 | 4904 | MRPL23 | 6150 | pp | -- | int.Intact: MI:0914(association) |
| RPS7 | 6201 | WDR46 | 9277 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS23 | 6228 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| YBX1 | 4904 | RPS24 | 6229 | pp | -- | int.Intact: MI:0914(association) |
| RPS16 | 6217 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| HSF1 | 3297 | MAPK8 | 5599 | pp | -- | int.I2D: HPRD, INNATEDB; int.HPRD: in vitro, in vivo |
| RPS3A | 6189 | RPS8 | 6202 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| MAPK14 | 1432 | ZNF672 | 79894 | pd | < | reg.ITFP.txt: no annot |
| COPA | 1314 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastMedium |
| NFKBIB | 4793 | SKP1 | 6500 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vitro |
| RPS6 | 6194 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| COPA | 1314 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid, HPRD, BCI; int.HPRD: in vivo |
| COPB1 | 1315 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, Tarassov_PCA, YeastMedium; int.DIP: MI:0915(physical association) |
| COPA | 1314 | ATP6V1D | 51382 | pp | -- | int.I2D: YeastLow |
| RPS15 | 6209 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastMedium |
| RPS11 | 6205 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BCI, HPRD, IntAct, MINT; int.Mint: MI:0915(physical association); int.HPRD: in vitro |
| RPS3A | 6189 | RPS15 | 6209 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS4X | 6191 | RPS8 | 6202 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS4X | 6191 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS15A | 6210 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| HSF1 | 3297 | MRPS12 | 6183 | pd | <> | reg.ITFP.txt: no annot |
| RAN | 5901 | SPHK2 | 56848 | pp | -- | int.I2D: IntAct_Yeast, BIND_Yeast, MINT_Yeast, MIPS, YeastLow |
| CDC5L | 988 | YBX1 | 4904 | pd | <> | reg.ITFP.txt: no annot |
| YBX1 | 4904 | MRPS12 | 6183 | pd | <> | reg.ITFP.txt: no annot |
| RPS4X | 6191 | RPS15 | 6209 | pp | -- | int.I2D: INTEROLOG, YeastMedium |
| RPS11 | 6205 | RPS14 | 6208 | pp | -- | int.I2D: BioGrid_Yeast |
| CDC5L | 988 | COPA | 1314 | pd | <> | reg.ITFP.txt: no annot |
| COPB1 | 1315 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid |
| YBX1 | 4904 | RPAP2 | 79871 | pp | -- | int.I2D: BioGrid |
| ARCN1 | 372 | COPB1 | 1315 | pp | -- | int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, BIND, BIND_Yeast, HPRD, Krogan_Core, MIPS; int.HPRD: in vivo |
| RPS9 | 6203 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS15 | 6209 | RPS15A | 6210 | pp | -- | int.I2D: YeastMedium, INTEROLOG |
| RPS6 | 6194 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | RPS3A | 6189 | pp | -- | int.I2D: IntAct_Yeast |
| RPS16 | 6217 | RPS23 | 6228 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS20 | 6224 | pp | -- | int.I2D: YeastLow |
| RPS6 | 6194 | RPS7 | 6201 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | COPA | 1314 | pp | -- | int.I2D: BCI, BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh, HPRD, Krogan_Core; int.HPRD: in vivo |
| RPS6 | 6194 | RPS8 | 6202 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| RPS4X | 6191 | RPS6 | 6194 | pp | -- | int.I2D: BioGrid_Yeast |
| YBX1 | 4904 | RPS29 | 6235 | pp | -- | int.Intact: MI:0914(association) |
| CDC5L | 988 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid |
| COPA | 1314 | RPS27A | 6233 | pp | -- | int.I2D: YeastLow |
| MAPK8 | 5599 | WWOX | 51741 | pp | -- | int.I2D: MINT, HPRD; int.Mint: MI:0915(physical association); int.HPRD: in vivo, yeast 2-hybrid |
| RPS8 | 6202 | RPS15A | 6210 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS7 | 6201 | RPS8 | 6202 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS23 | 6228 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS16 | 6217 | RPS26 | 6231 | pp | -- | int.I2D: BioGrid_Yeast |
| RAN | 5901 | ATP6V1D | 51382 | pp | -- | int.I2D: YeastLow |
| RPS15A | 6210 | RPS27A | 6233 | pp | -- | int.I2D: YeastLow |
| ARCN1 | 372 | COPB2 | 9276 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| RAN | 5901 | SKP1 | 6500 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS8 | 6202 | MAP3K14 | 9020 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, MINT; int.Mint: MI:0915(physical association) |
| RPS11 | 6205 | RPS20 | 6224 | pp | -- | int.I2D: YeastMedium, INTEROLOG |
| COPB1 | 1315 | MAPK14 | 1432 | pp | -- | int.I2D: IntAct_Mouse, MINT_Mouse |
| RPS7 | 6201 | RPS16 | 6217 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | RPS15 | 6209 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS24 | 6229 | RPS27A | 6233 | pp | -- | int.I2D: YeastMedium |
| RPS8 | 6202 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS3A | 6189 | RPS6 | 6194 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
Related GO terms (600)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0022627 | cytosolic small ribosomal subunit | 3.59e-39 | 5.86e-35 | 7.285 | 19 | 21 | 39 |
| GO:0019058 | viral life cycle | 7.13e-35 | 1.16e-30 | 5.936 | 22 | 25 | 115 |
| GO:0019083 | viral transcription | 7.29e-32 | 1.19e-27 | 6.230 | 19 | 22 | 81 |
| GO:0006415 | translational termination | 3.33e-31 | 5.43e-27 | 6.127 | 19 | 22 | 87 |
| GO:0003735 | structural constituent of ribosome | 9.07e-31 | 1.48e-26 | 5.575 | 21 | 24 | 141 |
| GO:0006414 | translational elongation | 1.35e-30 | 2.21e-26 | 6.031 | 19 | 22 | 93 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1.39e-29 | 2.26e-25 | 5.870 | 19 | 22 | 104 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 9.16e-29 | 1.49e-24 | 5.737 | 19 | 22 | 114 |
| GO:0006413 | translational initiation | 1.55e-27 | 2.52e-23 | 5.537 | 19 | 27 | 131 |
| GO:0016071 | mRNA metabolic process | 2.16e-26 | 3.52e-22 | 4.913 | 21 | 29 | 223 |
| GO:0006412 | translation | 6.68e-26 | 1.09e-21 | 4.838 | 21 | 29 | 235 |
| GO:0016070 | RNA metabolic process | 1.95e-25 | 3.18e-21 | 4.766 | 21 | 29 | 247 |
| GO:0016032 | viral process | 2.06e-25 | 3.36e-21 | 3.946 | 26 | 37 | 540 |
| GO:0005829 | cytosol | 4.14e-23 | 6.75e-19 | 2.321 | 40 | 74 | 2562 |
| GO:0010467 | gene expression | 5.69e-19 | 9.29e-15 | 3.460 | 23 | 36 | 669 |
| GO:0044822 | poly(A) RNA binding | 7.78e-18 | 1.27e-13 | 2.948 | 26 | 42 | 1078 |
| GO:0015935 | small ribosomal subunit | 1.22e-16 | 1.98e-12 | 7.235 | 8 | 9 | 17 |
| GO:0044267 | cellular protein metabolic process | 2.01e-16 | 3.27e-12 | 3.619 | 19 | 29 | 495 |
| GO:0016020 | membrane | 8.44e-16 | 1.38e-11 | 2.410 | 29 | 48 | 1746 |
| GO:0005925 | focal adhesion | 2.46e-13 | 4.02e-09 | 3.698 | 15 | 23 | 370 |
| GO:0042274 | ribosomal small subunit biogenesis | 6.26e-13 | 1.02e-08 | 7.322 | 6 | 6 | 12 |
| GO:0070062 | extracellular vesicular exosome | 8.15e-11 | 1.33e-06 | 1.832 | 28 | 51 | 2516 |
| GO:0048205 | COPI coating of Golgi vesicle | 3.08e-10 | 5.02e-06 | 6.944 | 5 | 6 | 13 |
| GO:0030126 | COPI vesicle coat | 3.08e-10 | 5.02e-06 | 6.944 | 5 | 6 | 13 |
| GO:0005840 | ribosome | 5.71e-10 | 9.33e-06 | 5.247 | 7 | 10 | 59 |
| GO:0061024 | membrane organization | 6.32e-10 | 1.03e-05 | 4.302 | 9 | 11 | 146 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 1.23e-08 | 2.01e-04 | 6.000 | 5 | 6 | 25 |
| GO:0030529 | ribonucleoprotein complex | 6.05e-08 | 9.88e-04 | 4.296 | 7 | 8 | 114 |
| GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2.86e-07 | 4.67e-03 | 7.585 | 3 | 3 | 5 |
| GO:0006364 | rRNA processing | 5.14e-07 | 8.40e-03 | 4.322 | 6 | 8 | 96 |
| GO:0000028 | ribosomal small subunit assembly | 9.97e-07 | 1.63e-02 | 7.100 | 3 | 3 | 7 |
| GO:0005515 | protein binding | 1.66e-06 | 2.72e-02 | 0.911 | 36 | 87 | 6127 |
| GO:0034146 | toll-like receptor 5 signaling pathway | 1.75e-06 | 2.85e-02 | 4.622 | 5 | 5 | 65 |
| GO:0034166 | toll-like receptor 10 signaling pathway | 1.75e-06 | 2.85e-02 | 4.622 | 5 | 5 | 65 |
| GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway | 2.72e-06 | 4.43e-02 | 4.494 | 5 | 5 | 71 |
| GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway | 2.72e-06 | 4.43e-02 | 4.494 | 5 | 5 | 71 |
| GO:0034162 | toll-like receptor 9 signaling pathway | 2.91e-06 | 4.75e-02 | 4.474 | 5 | 5 | 72 |
| GO:0034134 | toll-like receptor 2 signaling pathway | 3.12e-06 | 5.09e-02 | 4.454 | 5 | 5 | 73 |
| GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 3.81e-06 | 6.22e-02 | 4.396 | 5 | 5 | 76 |
| GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 4.33e-06 | 7.07e-02 | 4.359 | 5 | 5 | 78 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 4.91e-06 | 8.01e-02 | 4.322 | 5 | 5 | 80 |
| GO:0034138 | toll-like receptor 3 signaling pathway | 4.91e-06 | 8.01e-02 | 4.322 | 5 | 5 | 80 |
| GO:0005737 | cytoplasm | 1.03e-05 | 1.68e-01 | 1.120 | 27 | 65 | 3976 |
| GO:0034142 | toll-like receptor 4 signaling pathway | 1.20e-05 | 1.96e-01 | 4.059 | 5 | 5 | 96 |
| GO:0075733 | intracellular transport of virus | 1.90e-05 | 3.09e-01 | 5.820 | 3 | 3 | 17 |
| GO:0006891 | intra-Golgi vesicle-mediated transport | 1.90e-05 | 3.09e-01 | 5.820 | 3 | 3 | 17 |
| GO:0002224 | toll-like receptor signaling pathway | 2.23e-05 | 3.64e-01 | 3.876 | 5 | 5 | 109 |
| GO:0051403 | stress-activated MAPK cascade | 2.38e-05 | 3.89e-01 | 4.567 | 4 | 4 | 54 |
| GO:0000056 | ribosomal small subunit export from nucleus | 2.87e-05 | 4.68e-01 | 7.737 | 2 | 2 | 3 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 4.25e-05 | 6.93e-01 | 5.448 | 3 | 5 | 22 |
| GO:0043066 | negative regulation of apoptotic process | 5.39e-05 | 8.79e-01 | 2.564 | 8 | 14 | 433 |
| GO:0030490 | maturation of SSU-rRNA | 5.72e-05 | 9.34e-01 | 7.322 | 2 | 2 | 4 |
| GO:0019843 | rRNA binding | 7.97e-05 | 1.00e+00 | 5.152 | 3 | 4 | 27 |
| GO:0033119 | negative regulation of RNA splicing | 9.52e-05 | 1.00e+00 | 7.000 | 2 | 2 | 5 |
| GO:0003723 | RNA binding | 1.10e-04 | 1.00e+00 | 2.658 | 7 | 10 | 355 |
| GO:0005730 | nucleolus | 1.31e-04 | 1.00e+00 | 1.511 | 15 | 36 | 1684 |
| GO:0045182 | translation regulator activity | 1.42e-04 | 1.00e+00 | 6.737 | 2 | 2 | 6 |
| GO:0002181 | cytoplasmic translation | 1.42e-04 | 1.00e+00 | 6.737 | 2 | 2 | 6 |
| GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 1.91e-04 | 1.00e+00 | 4.737 | 3 | 3 | 36 |
| GO:0006886 | intracellular protein transport | 2.00e-04 | 1.00e+00 | 3.209 | 5 | 6 | 173 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 2.82e-04 | 1.00e+00 | 4.549 | 3 | 4 | 41 |
| GO:0007254 | JNK cascade | 5.39e-04 | 1.00e+00 | 4.235 | 3 | 3 | 51 |
| GO:0019082 | viral protein processing | 6.19e-04 | 1.00e+00 | 5.737 | 2 | 2 | 12 |
| GO:0032479 | regulation of type I interferon production | 7.30e-04 | 1.00e+00 | 5.622 | 2 | 2 | 13 |
| GO:0071398 | cellular response to fatty acid | 7.30e-04 | 1.00e+00 | 5.622 | 2 | 2 | 13 |
| GO:0016197 | endosomal transport | 7.86e-04 | 1.00e+00 | 4.049 | 3 | 3 | 58 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 8.75e-04 | 1.00e+00 | 3.224 | 4 | 6 | 137 |
| GO:0032481 | positive regulation of type I interferon production | 9.11e-04 | 1.00e+00 | 3.976 | 3 | 3 | 61 |
| GO:0031295 | T cell costimulation | 1.20e-03 | 1.00e+00 | 3.841 | 3 | 3 | 67 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.22e-03 | 1.00e+00 | 3.093 | 4 | 11 | 150 |
| GO:0019068 | virion assembly | 1.26e-03 | 1.00e+00 | 5.235 | 2 | 2 | 17 |
| GO:0018105 | peptidyl-serine phosphorylation | 1.30e-03 | 1.00e+00 | 3.798 | 3 | 5 | 69 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1.36e-03 | 1.00e+00 | 3.778 | 3 | 8 | 70 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 1.58e-03 | 1.00e+00 | 2.551 | 5 | 5 | 273 |
| GO:0043005 | neuron projection | 1.59e-03 | 1.00e+00 | 2.991 | 4 | 6 | 161 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1.59e-03 | 1.00e+00 | 3.698 | 3 | 8 | 74 |
| GO:0043065 | positive regulation of apoptotic process | 1.61e-03 | 1.00e+00 | 2.546 | 5 | 6 | 274 |
| GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity | 1.75e-03 | 1.00e+00 | 5.000 | 2 | 2 | 20 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 1.92e-03 | 1.00e+00 | 3.603 | 3 | 8 | 79 |
| GO:0071222 | cellular response to lipopolysaccharide | 2.07e-03 | 1.00e+00 | 3.567 | 3 | 4 | 81 |
| GO:0007220 | Notch receptor processing | 2.12e-03 | 1.00e+00 | 4.863 | 2 | 2 | 22 |
| GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 2.74e-03 | 1.00e+00 | 4.678 | 2 | 2 | 25 |
| GO:0005844 | polysome | 2.74e-03 | 1.00e+00 | 4.678 | 2 | 2 | 25 |
| GO:0000187 | activation of MAPK activity | 2.79e-03 | 1.00e+00 | 3.415 | 3 | 3 | 90 |
| GO:0045087 | innate immune response | 2.88e-03 | 1.00e+00 | 1.863 | 7 | 11 | 616 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 2.96e-03 | 1.00e+00 | 2.745 | 4 | 5 | 191 |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2.97e-03 | 1.00e+00 | 4.622 | 2 | 2 | 26 |
| GO:0005978 | glycogen biosynthetic process | 2.97e-03 | 1.00e+00 | 4.622 | 2 | 2 | 26 |
| GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 2.97e-03 | 1.00e+00 | 4.622 | 2 | 2 | 26 |
| GO:2001286 | regulation of caveolin-mediated endocytosis | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0071393 | cellular response to progesterone stimulus | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0033176 | proton-transporting V-type ATPase complex | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0004673 | protein histidine kinase activity | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0006669 | sphinganine-1-phosphate biosynthetic process | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0022605 | oogenesis stage | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0018106 | peptidyl-histidine phosphorylation | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0021691 | cerebellar Purkinje cell layer maturation | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0038066 | p38MAPK cascade | 3.12e-03 | 1.00e+00 | 8.322 | 1 | 1 | 1 |
| GO:0051149 | positive regulation of muscle cell differentiation | 3.20e-03 | 1.00e+00 | 4.567 | 2 | 2 | 27 |
| GO:0001649 | osteoblast differentiation | 3.25e-03 | 1.00e+00 | 3.337 | 3 | 3 | 95 |
| GO:0071456 | cellular response to hypoxia | 3.55e-03 | 1.00e+00 | 3.292 | 3 | 3 | 98 |
| GO:0032480 | negative regulation of type I interferon production | 4.47e-03 | 1.00e+00 | 4.322 | 2 | 2 | 32 |
| GO:0042692 | muscle cell differentiation | 5.04e-03 | 1.00e+00 | 4.235 | 2 | 2 | 34 |
| GO:0034332 | adherens junction organization | 5.64e-03 | 1.00e+00 | 4.152 | 2 | 4 | 36 |
| GO:0097190 | apoptotic signaling pathway | 5.68e-03 | 1.00e+00 | 3.049 | 3 | 3 | 116 |
| GO:0000209 | protein polyubiquitination | 5.68e-03 | 1.00e+00 | 3.049 | 3 | 7 | 116 |
| GO:0006006 | glucose metabolic process | 6.10e-03 | 1.00e+00 | 3.012 | 3 | 4 | 119 |
| GO:0008481 | sphinganine kinase activity | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0071987 | WD40-repeat domain binding | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0060661 | submandibular salivary gland formation | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0002762 | negative regulation of myeloid leukocyte differentiation | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0017050 | D-erythro-sphingosine kinase activity | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0071338 | positive regulation of hair follicle cell proliferation | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0072422 | signal transduction involved in DNA damage checkpoint | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0010632 | regulation of epithelial cell migration | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0090135 | actin filament branching | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 6.24e-03 | 1.00e+00 | 7.322 | 1 | 1 | 2 |
| GO:0030218 | erythrocyte differentiation | 6.26e-03 | 1.00e+00 | 4.074 | 2 | 2 | 38 |
| GO:0003713 | transcription coactivator activity | 6.54e-03 | 1.00e+00 | 2.421 | 4 | 6 | 239 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 6.99e-03 | 1.00e+00 | 2.941 | 3 | 3 | 125 |
| GO:0007219 | Notch signaling pathway | 6.99e-03 | 1.00e+00 | 2.941 | 3 | 4 | 125 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 7.78e-03 | 1.00e+00 | 2.885 | 3 | 4 | 130 |
| GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 7.97e-03 | 1.00e+00 | 3.896 | 2 | 2 | 43 |
| GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0003161 | cardiac conduction system development | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0051154 | negative regulation of striated muscle cell differentiation | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0090045 | positive regulation of deacetylase activity | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0035033 | histone deacetylase regulator activity | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:2000017 | positive regulation of determination of dorsal identity | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0051683 | establishment of Golgi localization | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 2 | 3 |
| GO:0006670 | sphingosine metabolic process | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0090400 | stress-induced premature senescence | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0004705 | JUN kinase activity | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 9.35e-03 | 1.00e+00 | 6.737 | 1 | 1 | 3 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 9.77e-03 | 1.00e+00 | 1.705 | 6 | 9 | 589 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 1.11e-02 | 1.00e+00 | 2.698 | 3 | 3 | 148 |
| GO:0010628 | positive regulation of gene expression | 1.13e-02 | 1.00e+00 | 2.688 | 3 | 4 | 149 |
| GO:0042981 | regulation of apoptotic process | 1.17e-02 | 1.00e+00 | 2.669 | 3 | 7 | 151 |
| GO:0030686 | 90S preribosome | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0034191 | apolipoprotein A-I receptor binding | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0007000 | nucleolus organization | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 2 | 4 |
| GO:0030666 | endocytic vesicle membrane | 1.24e-02 | 1.00e+00 | 3.567 | 2 | 2 | 54 |
| GO:0009301 | snRNA transcription | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0032463 | negative regulation of protein homooligomerization | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0051902 | negative regulation of mitochondrial depolarization | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0060020 | Bergmann glial cell differentiation | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0031062 | positive regulation of histone methylation | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0060684 | epithelial-mesenchymal cell signaling | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0070851 | growth factor receptor binding | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0051835 | positive regulation of synapse structural plasticity | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0090231 | regulation of spindle checkpoint | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0072384 | organelle transport along microtubule | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 2 | 4 |
| GO:0007258 | JUN phosphorylation | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0048664 | neuron fate determination | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0033503 | HULC complex | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0045682 | regulation of epidermis development | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0031063 | regulation of histone deacetylation | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0031467 | Cul7-RING ubiquitin ligase complex | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0000055 | ribosomal large subunit export from nucleus | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0033625 | positive regulation of integrin activation | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0009991 | response to extracellular stimulus | 1.24e-02 | 1.00e+00 | 6.322 | 1 | 1 | 4 |
| GO:0005634 | nucleus | 1.34e-02 | 1.00e+00 | 0.608 | 23 | 66 | 4828 |
| GO:0005198 | structural molecule activity | 1.34e-02 | 1.00e+00 | 2.594 | 3 | 5 | 159 |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | 1.34e-02 | 1.00e+00 | 2.594 | 3 | 3 | 159 |
| GO:2000641 | regulation of early endosome to late endosome transport | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0036336 | dendritic cell migration | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0035088 | establishment or maintenance of apical/basal cell polarity | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0051385 | response to mineralocorticoid | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:1902188 | positive regulation of viral release from host cell | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0097300 | programmed necrotic cell death | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0031256 | leading edge membrane | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0008420 | CTD phosphatase activity | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0051525 | NFAT protein binding | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:1901741 | positive regulation of myoblast fusion | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0071803 | positive regulation of podosome assembly | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0046834 | lipid phosphorylation | 1.55e-02 | 1.00e+00 | 6.000 | 1 | 1 | 5 |
| GO:0038095 | Fc-epsilon receptor signaling pathway | 1.63e-02 | 1.00e+00 | 2.489 | 3 | 3 | 171 |
| GO:0006417 | regulation of translation | 1.66e-02 | 1.00e+00 | 3.345 | 2 | 4 | 63 |
| GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 1.71e-02 | 1.00e+00 | 3.322 | 2 | 2 | 64 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 1.76e-02 | 1.00e+00 | 3.300 | 2 | 6 | 65 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1.76e-02 | 1.00e+00 | 3.300 | 2 | 6 | 65 |
| GO:0019901 | protein kinase binding | 1.76e-02 | 1.00e+00 | 2.000 | 4 | 9 | 320 |
| GO:0005882 | intermediate filament | 1.81e-02 | 1.00e+00 | 3.278 | 2 | 3 | 66 |
| GO:0071260 | cellular response to mechanical stimulus | 1.81e-02 | 1.00e+00 | 3.278 | 2 | 3 | 66 |
| GO:0050847 | progesterone receptor signaling pathway | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0007143 | female meiotic division | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0002309 | T cell proliferation involved in immune response | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0004704 | NF-kappaB-inducing kinase activity | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0045056 | transcytosis | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0060789 | hair follicle placode formation | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0048554 | positive regulation of metalloenzyme activity | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0050792 | regulation of viral process | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0019215 | intermediate filament binding | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0000974 | Prp19 complex | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 1 | 6 |
| GO:0006924 | activation-induced cell death of T cells | 1.86e-02 | 1.00e+00 | 5.737 | 1 | 2 | 6 |
| GO:0005654 | nucleoplasm | 1.94e-02 | 1.00e+00 | 1.225 | 8 | 26 | 1095 |
| GO:0003697 | single-stranded DNA binding | 1.97e-02 | 1.00e+00 | 3.213 | 2 | 4 | 69 |
| GO:0003682 | chromatin binding | 2.02e-02 | 1.00e+00 | 1.938 | 4 | 4 | 334 |
| GO:0010447 | response to acidic pH | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0009142 | nucleoside triphosphate biosynthetic process | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0060136 | embryonic process involved in female pregnancy | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0043497 | regulation of protein heterodimerization activity | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0050658 | RNA transport | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0048027 | mRNA 5'-UTR binding | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0007097 | nuclear migration | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 2 | 7 |
| GO:0061512 | protein localization to cilium | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0003334 | keratinocyte development | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0034101 | erythrocyte homeostasis | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0010907 | positive regulation of glucose metabolic process | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0030157 | pancreatic juice secretion | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0000930 | gamma-tubulin complex | 2.17e-02 | 1.00e+00 | 5.515 | 1 | 1 | 7 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2.18e-02 | 1.00e+00 | 3.132 | 2 | 6 | 73 |
| GO:0003729 | mRNA binding | 2.18e-02 | 1.00e+00 | 3.132 | 2 | 3 | 73 |
| GO:0007265 | Ras protein signal transduction | 2.30e-02 | 1.00e+00 | 3.093 | 2 | 3 | 75 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 2.41e-02 | 1.00e+00 | 3.055 | 2 | 6 | 77 |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 2.41e-02 | 1.00e+00 | 3.055 | 2 | 4 | 77 |
| GO:0007229 | integrin-mediated signaling pathway | 2.47e-02 | 1.00e+00 | 3.037 | 2 | 2 | 78 |
| GO:0039702 | viral budding via host ESCRT complex | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0006183 | GTP biosynthetic process | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0043114 | regulation of vascular permeability | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0045124 | regulation of bone resorption | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0051489 | regulation of filopodium assembly | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0033523 | histone H2B ubiquitination | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0007172 | signal complex assembly | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0010831 | positive regulation of myotube differentiation | 2.47e-02 | 1.00e+00 | 5.322 | 1 | 1 | 8 |
| GO:0010629 | negative regulation of gene expression | 2.59e-02 | 1.00e+00 | 3.000 | 2 | 2 | 80 |
| GO:0016591 | DNA-directed RNA polymerase II, holoenzyme | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0038061 | NIK/NF-kappaB signaling | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0071732 | cellular response to nitric oxide | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0006241 | CTP biosynthetic process | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0014075 | response to amine | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0006228 | UTP biosynthetic process | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0090136 | epithelial cell-cell adhesion | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 2 | 9 |
| GO:0047497 | mitochondrion transport along microtubule | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0046628 | positive regulation of insulin receptor signaling pathway | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0032495 | response to muramyl dipeptide | 2.78e-02 | 1.00e+00 | 5.152 | 1 | 1 | 9 |
| GO:0045618 | positive regulation of keratinocyte differentiation | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 1 | 10 |
| GO:0032040 | small-subunit processome | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 1 | 10 |
| GO:0070307 | lens fiber cell development | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 1 | 10 |
| GO:0005798 | Golgi-associated vesicle | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 1 | 10 |
| GO:0031274 | positive regulation of pseudopodium assembly | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 2 | 10 |
| GO:0022407 | regulation of cell-cell adhesion | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 1 | 10 |
| GO:0060047 | heart contraction | 3.08e-02 | 1.00e+00 | 5.000 | 1 | 1 | 10 |
| GO:0050852 | T cell receptor signaling pathway | 3.09e-02 | 1.00e+00 | 2.863 | 2 | 2 | 88 |
| GO:0006915 | apoptotic process | 3.23e-02 | 1.00e+00 | 1.487 | 5 | 9 | 571 |
| GO:0000922 | spindle pole | 3.29e-02 | 1.00e+00 | 2.814 | 2 | 5 | 91 |
| GO:0006928 | cellular component movement | 3.35e-02 | 1.00e+00 | 2.798 | 2 | 4 | 92 |
| GO:0008284 | positive regulation of cell proliferation | 3.38e-02 | 1.00e+00 | 1.707 | 4 | 7 | 392 |
| GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0051895 | negative regulation of focal adhesion assembly | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0045120 | pronucleus | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:2000573 | positive regulation of DNA biosynthetic process | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 2 | 11 |
| GO:0051272 | positive regulation of cellular component movement | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0045663 | positive regulation of myoblast differentiation | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0045109 | intermediate filament organization | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0060065 | uterus development | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0019395 | fatty acid oxidation | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0004707 | MAP kinase activity | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0010390 | histone monoubiquitination | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0035518 | histone H2A monoubiquitination | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 2 | 11 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0014002 | astrocyte development | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0042770 | signal transduction in response to DNA damage | 3.39e-02 | 1.00e+00 | 4.863 | 1 | 1 | 11 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 3.49e-02 | 1.00e+00 | 2.767 | 2 | 7 | 94 |
| GO:0000278 | mitotic cell cycle | 3.54e-02 | 1.00e+00 | 1.685 | 4 | 16 | 398 |
| GO:0007067 | mitotic nuclear division | 3.54e-02 | 1.00e+00 | 2.055 | 3 | 7 | 231 |
| GO:0061136 | regulation of proteasomal protein catabolic process | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 1 | 12 |
| GO:0007051 | spindle organization | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 1 | 12 |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 1 | 12 |
| GO:0051146 | striated muscle cell differentiation | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 2 | 12 |
| GO:0043149 | stress fiber assembly | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 2 | 12 |
| GO:1903543 | positive regulation of exosomal secretion | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 1 | 12 |
| GO:1901214 | regulation of neuron death | 3.69e-02 | 1.00e+00 | 4.737 | 1 | 1 | 12 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 3.98e-02 | 1.00e+00 | 1.243 | 6 | 11 | 811 |
| GO:0043488 | regulation of mRNA stability | 3.99e-02 | 1.00e+00 | 4.622 | 1 | 1 | 13 |
| GO:0031929 | TOR signaling | 3.99e-02 | 1.00e+00 | 4.622 | 1 | 1 | 13 |
| GO:0004708 | MAP kinase kinase activity | 3.99e-02 | 1.00e+00 | 4.622 | 1 | 1 | 13 |
| GO:0005662 | DNA replication factor A complex | 3.99e-02 | 1.00e+00 | 4.622 | 1 | 1 | 13 |
| GO:0060444 | branching involved in mammary gland duct morphogenesis | 3.99e-02 | 1.00e+00 | 4.622 | 1 | 1 | 13 |
| GO:0004143 | diacylglycerol kinase activity | 3.99e-02 | 1.00e+00 | 4.622 | 1 | 1 | 13 |
| GO:0035371 | microtubule plus-end | 4.29e-02 | 1.00e+00 | 4.515 | 1 | 1 | 14 |
| GO:0031333 | negative regulation of protein complex assembly | 4.29e-02 | 1.00e+00 | 4.515 | 1 | 1 | 14 |
| GO:0050662 | coenzyme binding | 4.29e-02 | 1.00e+00 | 4.515 | 1 | 1 | 14 |
| GO:0017016 | Ras GTPase binding | 4.29e-02 | 1.00e+00 | 4.515 | 1 | 1 | 14 |
| GO:0031996 | thioesterase binding | 4.29e-02 | 1.00e+00 | 4.515 | 1 | 1 | 14 |
| GO:0006165 | nucleoside diphosphate phosphorylation | 4.29e-02 | 1.00e+00 | 4.515 | 1 | 1 | 14 |
| GO:0005739 | mitochondrion | 4.29e-02 | 1.00e+00 | 1.099 | 7 | 10 | 1046 |
| GO:0048477 | oogenesis | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0030131 | clathrin adaptor complex | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0030225 | macrophage differentiation | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0003951 | NAD+ kinase activity | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0004550 | nucleoside diphosphate kinase activity | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0048821 | erythrocyte development | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 2 | 15 |
| GO:0042307 | positive regulation of protein import into nucleus | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 2 | 15 |
| GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0031369 | translation initiation factor binding | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 2 | 15 |
| GO:0051233 | spindle midzone | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 2 | 15 |
| GO:0005762 | mitochondrial large ribosomal subunit | 4.59e-02 | 1.00e+00 | 4.415 | 1 | 1 | 15 |
| GO:0005815 | microtubule organizing center | 4.63e-02 | 1.00e+00 | 2.541 | 2 | 4 | 110 |
| GO:0014911 | positive regulation of smooth muscle cell migration | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 16 |
| GO:0048037 | cofactor binding | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 16 |
| GO:0042176 | regulation of protein catabolic process | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 4 | 16 |
| GO:2000811 | negative regulation of anoikis | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 16 |
| GO:0005212 | structural constituent of eye lens | 4.89e-02 | 1.00e+00 | 4.322 | 1 | 1 | 16 |
| GO:0002548 | monocyte chemotaxis | 5.18e-02 | 1.00e+00 | 4.235 | 1 | 1 | 17 |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | 5.18e-02 | 1.00e+00 | 4.235 | 1 | 1 | 17 |
| GO:0030742 | GTP-dependent protein binding | 5.18e-02 | 1.00e+00 | 4.235 | 1 | 1 | 17 |
| GO:0070064 | proline-rich region binding | 5.48e-02 | 1.00e+00 | 4.152 | 1 | 1 | 18 |
| GO:0004709 | MAP kinase kinase kinase activity | 5.48e-02 | 1.00e+00 | 4.152 | 1 | 1 | 18 |
| GO:0031954 | positive regulation of protein autophosphorylation | 5.48e-02 | 1.00e+00 | 4.152 | 1 | 1 | 18 |
| GO:0015949 | nucleobase-containing small molecule interconversion | 5.48e-02 | 1.00e+00 | 4.152 | 1 | 1 | 18 |
| GO:0043015 | gamma-tubulin binding | 5.48e-02 | 1.00e+00 | 4.152 | 1 | 2 | 18 |
| GO:0090316 | positive regulation of intracellular protein transport | 5.48e-02 | 1.00e+00 | 4.152 | 1 | 1 | 18 |
| GO:0006259 | DNA metabolic process | 5.78e-02 | 1.00e+00 | 4.074 | 1 | 1 | 19 |
| GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 5.78e-02 | 1.00e+00 | 4.074 | 1 | 1 | 19 |
| GO:0007088 | regulation of mitosis | 5.78e-02 | 1.00e+00 | 4.074 | 1 | 1 | 19 |
| GO:0017134 | fibroblast growth factor binding | 5.78e-02 | 1.00e+00 | 4.074 | 1 | 2 | 19 |
| GO:0045453 | bone resorption | 5.78e-02 | 1.00e+00 | 4.074 | 1 | 1 | 19 |
| GO:0031667 | response to nutrient levels | 5.78e-02 | 1.00e+00 | 4.074 | 1 | 1 | 19 |
| GO:0032148 | activation of protein kinase B activity | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 1 | 20 |
| GO:0001502 | cartilage condensation | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 1 | 20 |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 1 | 20 |
| GO:0043473 | pigmentation | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 2 | 20 |
| GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 1 | 20 |
| GO:0043393 | regulation of protein binding | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 2 | 20 |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 6.36e-02 | 1.00e+00 | 3.930 | 1 | 1 | 21 |
| GO:0045648 | positive regulation of erythrocyte differentiation | 6.36e-02 | 1.00e+00 | 3.930 | 1 | 1 | 21 |
| GO:0007369 | gastrulation | 6.36e-02 | 1.00e+00 | 3.930 | 1 | 1 | 21 |
| GO:0005689 | U12-type spliceosomal complex | 6.36e-02 | 1.00e+00 | 3.930 | 1 | 2 | 21 |
| GO:0046847 | filopodium assembly | 6.36e-02 | 1.00e+00 | 3.930 | 1 | 1 | 21 |
| GO:0051881 | regulation of mitochondrial membrane potential | 6.36e-02 | 1.00e+00 | 3.930 | 1 | 1 | 21 |
| GO:0009615 | response to virus | 6.40e-02 | 1.00e+00 | 2.278 | 2 | 4 | 132 |
| GO:0030316 | osteoclast differentiation | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:0046686 | response to cadmium ion | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:0033574 | response to testosterone | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:0001106 | RNA polymerase II transcription corepressor activity | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:0031435 | mitogen-activated protein kinase kinase kinase binding | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:0007052 | mitotic spindle organization | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 22 |
| GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 1 | 23 |
| GO:0071944 | cell periphery | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 1 | 23 |
| GO:0045787 | positive regulation of cell cycle | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 2 | 23 |
| GO:0002040 | sprouting angiogenesis | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 1 | 23 |
| GO:0051017 | actin filament bundle assembly | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 1 | 23 |
| GO:0001892 | embryonic placenta development | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 1 | 23 |
| GO:0051297 | centrosome organization | 6.95e-02 | 1.00e+00 | 3.798 | 1 | 2 | 23 |
| GO:0005761 | mitochondrial ribosome | 7.24e-02 | 1.00e+00 | 3.737 | 1 | 1 | 24 |
| GO:0007163 | establishment or maintenance of cell polarity | 7.24e-02 | 1.00e+00 | 3.737 | 1 | 2 | 24 |
| GO:0051602 | response to electrical stimulus | 7.24e-02 | 1.00e+00 | 3.737 | 1 | 1 | 24 |
| GO:2000379 | positive regulation of reactive oxygen species metabolic process | 7.24e-02 | 1.00e+00 | 3.737 | 1 | 1 | 24 |
| GO:0008286 | insulin receptor signaling pathway | 7.45e-02 | 1.00e+00 | 2.152 | 2 | 4 | 144 |
| GO:0006611 | protein export from nucleus | 7.53e-02 | 1.00e+00 | 3.678 | 1 | 1 | 25 |
| GO:0031519 | PcG protein complex | 7.53e-02 | 1.00e+00 | 3.678 | 1 | 1 | 25 |
| GO:0071479 | cellular response to ionizing radiation | 7.53e-02 | 1.00e+00 | 3.678 | 1 | 2 | 25 |
| GO:0050715 | positive regulation of cytokine secretion | 7.53e-02 | 1.00e+00 | 3.678 | 1 | 1 | 25 |
| GO:0048705 | skeletal system morphogenesis | 7.53e-02 | 1.00e+00 | 3.678 | 1 | 1 | 25 |
| GO:0045859 | regulation of protein kinase activity | 7.82e-02 | 1.00e+00 | 3.622 | 1 | 1 | 26 |
| GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 7.82e-02 | 1.00e+00 | 3.622 | 1 | 1 | 26 |
| GO:0030148 | sphingolipid biosynthetic process | 7.82e-02 | 1.00e+00 | 3.622 | 1 | 1 | 26 |
| GO:0032720 | negative regulation of tumor necrosis factor production | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 1 | 27 |
| GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 1 | 27 |
| GO:0031424 | keratinization | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 1 | 27 |
| GO:0001103 | RNA polymerase II repressing transcription factor binding | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 1 | 27 |
| GO:0030331 | estrogen receptor binding | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 1 | 27 |
| GO:0045184 | establishment of protein localization | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 2 | 27 |
| GO:0031069 | hair follicle morphogenesis | 8.11e-02 | 1.00e+00 | 3.567 | 1 | 1 | 27 |
| GO:0032467 | positive regulation of cytokinesis | 8.40e-02 | 1.00e+00 | 3.515 | 1 | 1 | 28 |
| GO:0000077 | DNA damage checkpoint | 8.40e-02 | 1.00e+00 | 3.515 | 1 | 1 | 28 |
| GO:0042626 | ATPase activity, coupled to transmembrane movement of substances | 8.40e-02 | 1.00e+00 | 3.515 | 1 | 1 | 28 |
| GO:0043231 | intracellular membrane-bounded organelle | 8.51e-02 | 1.00e+00 | 1.532 | 3 | 3 | 332 |
| GO:0010008 | endosome membrane | 8.64e-02 | 1.00e+00 | 2.027 | 2 | 2 | 157 |
| GO:0031252 | cell leading edge | 8.68e-02 | 1.00e+00 | 3.464 | 1 | 3 | 29 |
| GO:0019005 | SCF ubiquitin ligase complex | 8.68e-02 | 1.00e+00 | 3.464 | 1 | 1 | 29 |
| GO:0003730 | mRNA 3'-UTR binding | 8.68e-02 | 1.00e+00 | 3.464 | 1 | 1 | 29 |
| GO:0034605 | cellular response to heat | 8.68e-02 | 1.00e+00 | 3.464 | 1 | 1 | 29 |
| GO:0072686 | mitotic spindle | 8.68e-02 | 1.00e+00 | 3.464 | 1 | 1 | 29 |
| GO:0031663 | lipopolysaccharide-mediated signaling pathway | 8.68e-02 | 1.00e+00 | 3.464 | 1 | 1 | 29 |
| GO:0005813 | centrosome | 8.93e-02 | 1.00e+00 | 1.502 | 3 | 9 | 339 |
| GO:0010977 | negative regulation of neuron projection development | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 2 | 30 |
| GO:0051262 | protein tetramerization | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 1 | 30 |
| GO:0046875 | ephrin receptor binding | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 2 | 30 |
| GO:0042169 | SH2 domain binding | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 1 | 30 |
| GO:0031647 | regulation of protein stability | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 1 | 30 |
| GO:0040018 | positive regulation of multicellular organism growth | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 1 | 30 |
| GO:0010494 | cytoplasmic stress granule | 8.97e-02 | 1.00e+00 | 3.415 | 1 | 2 | 30 |
| GO:0008022 | protein C-terminus binding | 9.01e-02 | 1.00e+00 | 1.991 | 2 | 4 | 161 |
| GO:0070555 | response to interleukin-1 | 9.25e-02 | 1.00e+00 | 3.368 | 1 | 1 | 31 |
| GO:0007093 | mitotic cell cycle checkpoint | 9.25e-02 | 1.00e+00 | 3.368 | 1 | 2 | 31 |
| GO:0000398 | mRNA splicing, via spliceosome | 9.40e-02 | 1.00e+00 | 1.956 | 2 | 2 | 165 |
| GO:0033572 | transferrin transport | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 1 | 32 |
| GO:0002062 | chondrocyte differentiation | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 1 | 32 |
| GO:0015992 | proton transport | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 1 | 32 |
| GO:0051219 | phosphoprotein binding | 9.54e-02 | 1.00e+00 | 3.322 | 1 | 4 | 32 |
| GO:0034220 | ion transmembrane transport | 9.59e-02 | 1.00e+00 | 1.938 | 2 | 2 | 167 |
| GO:0032091 | negative regulation of protein binding | 9.82e-02 | 1.00e+00 | 3.278 | 1 | 1 | 33 |
| GO:0005158 | insulin receptor binding | 9.82e-02 | 1.00e+00 | 3.278 | 1 | 2 | 33 |
| GO:0033077 | T cell differentiation in thymus | 9.82e-02 | 1.00e+00 | 3.278 | 1 | 1 | 33 |
| GO:0048812 | neuron projection morphogenesis | 9.82e-02 | 1.00e+00 | 3.278 | 1 | 1 | 33 |
| GO:0001890 | placenta development | 1.01e-01 | 1.00e+00 | 3.235 | 1 | 1 | 34 |
| GO:0097110 | scaffold protein binding | 1.01e-01 | 1.00e+00 | 3.235 | 1 | 1 | 34 |
| GO:0051701 | interaction with host | 1.01e-01 | 1.00e+00 | 3.235 | 1 | 1 | 34 |
| GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway | 1.04e-01 | 1.00e+00 | 3.193 | 1 | 1 | 35 |
| GO:0071333 | cellular response to glucose stimulus | 1.04e-01 | 1.00e+00 | 3.193 | 1 | 2 | 35 |
| GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 1.04e-01 | 1.00e+00 | 3.193 | 1 | 1 | 35 |
| GO:0030178 | negative regulation of Wnt signaling pathway | 1.07e-01 | 1.00e+00 | 3.152 | 1 | 1 | 36 |
| GO:0004672 | protein kinase activity | 1.07e-01 | 1.00e+00 | 1.846 | 2 | 4 | 178 |
| GO:0071560 | cellular response to transforming growth factor beta stimulus | 1.07e-01 | 1.00e+00 | 3.152 | 1 | 1 | 36 |
| GO:0016301 | kinase activity | 1.09e-01 | 1.00e+00 | 3.113 | 1 | 2 | 37 |
| GO:0018107 | peptidyl-threonine phosphorylation | 1.09e-01 | 1.00e+00 | 3.113 | 1 | 1 | 37 |
| GO:0032880 | regulation of protein localization | 1.09e-01 | 1.00e+00 | 3.113 | 1 | 1 | 37 |
| GO:0019904 | protein domain specific binding | 1.10e-01 | 1.00e+00 | 1.822 | 2 | 3 | 181 |
| GO:0001568 | blood vessel development | 1.12e-01 | 1.00e+00 | 3.074 | 1 | 2 | 38 |
| GO:0050681 | androgen receptor binding | 1.12e-01 | 1.00e+00 | 3.074 | 1 | 2 | 38 |
| GO:0030049 | muscle filament sliding | 1.12e-01 | 1.00e+00 | 3.074 | 1 | 2 | 38 |
| GO:0097191 | extrinsic apoptotic signaling pathway | 1.12e-01 | 1.00e+00 | 3.074 | 1 | 3 | 38 |
| GO:0045740 | positive regulation of DNA replication | 1.12e-01 | 1.00e+00 | 3.074 | 1 | 1 | 38 |
| GO:0090382 | phagosome maturation | 1.12e-01 | 1.00e+00 | 3.074 | 1 | 1 | 38 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.13e-01 | 1.00e+00 | 1.798 | 2 | 5 | 184 |
| GO:0051781 | positive regulation of cell division | 1.15e-01 | 1.00e+00 | 3.037 | 1 | 1 | 39 |
| GO:0032092 | positive regulation of protein binding | 1.15e-01 | 1.00e+00 | 3.037 | 1 | 1 | 39 |
| GO:0021766 | hippocampus development | 1.15e-01 | 1.00e+00 | 3.037 | 1 | 3 | 39 |
| GO:0006351 | transcription, DNA-templated | 1.17e-01 | 1.00e+00 | 0.692 | 8 | 17 | 1585 |
| GO:0042542 | response to hydrogen peroxide | 1.18e-01 | 1.00e+00 | 3.000 | 1 | 1 | 40 |
| GO:0070301 | cellular response to hydrogen peroxide | 1.21e-01 | 1.00e+00 | 2.964 | 1 | 1 | 41 |
| GO:0030521 | androgen receptor signaling pathway | 1.21e-01 | 1.00e+00 | 2.964 | 1 | 3 | 41 |
| GO:0045785 | positive regulation of cell adhesion | 1.21e-01 | 1.00e+00 | 2.964 | 1 | 1 | 41 |
| GO:0007519 | skeletal muscle tissue development | 1.21e-01 | 1.00e+00 | 2.964 | 1 | 1 | 41 |
| GO:0005902 | microvillus | 1.23e-01 | 1.00e+00 | 2.930 | 1 | 1 | 42 |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | 1.23e-01 | 1.00e+00 | 2.930 | 1 | 1 | 42 |
| GO:0007286 | spermatid development | 1.29e-01 | 1.00e+00 | 2.863 | 1 | 1 | 44 |
| GO:0005080 | protein kinase C binding | 1.29e-01 | 1.00e+00 | 2.863 | 1 | 1 | 44 |
| GO:0034613 | cellular protein localization | 1.29e-01 | 1.00e+00 | 2.863 | 1 | 2 | 44 |
| GO:0009411 | response to UV | 1.32e-01 | 1.00e+00 | 2.830 | 1 | 2 | 45 |
| GO:0003924 | GTPase activity | 1.32e-01 | 1.00e+00 | 1.657 | 2 | 6 | 203 |
| GO:0051591 | response to cAMP | 1.32e-01 | 1.00e+00 | 2.830 | 1 | 1 | 45 |
| GO:0030168 | platelet activation | 1.34e-01 | 1.00e+00 | 1.643 | 2 | 4 | 205 |
| GO:0044297 | cell body | 1.34e-01 | 1.00e+00 | 2.798 | 1 | 1 | 46 |
| GO:0043525 | positive regulation of neuron apoptotic process | 1.34e-01 | 1.00e+00 | 2.798 | 1 | 2 | 46 |
| GO:0045727 | positive regulation of translation | 1.34e-01 | 1.00e+00 | 2.798 | 1 | 1 | 46 |
| GO:0021762 | substantia nigra development | 1.34e-01 | 1.00e+00 | 2.798 | 1 | 2 | 46 |
| GO:0043406 | positive regulation of MAP kinase activity | 1.37e-01 | 1.00e+00 | 2.767 | 1 | 1 | 47 |
| GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 1.37e-01 | 1.00e+00 | 2.767 | 1 | 1 | 47 |
| GO:0008344 | adult locomotory behavior | 1.37e-01 | 1.00e+00 | 2.767 | 1 | 2 | 47 |
| GO:0006950 | response to stress | 1.40e-01 | 1.00e+00 | 2.737 | 1 | 1 | 48 |
| GO:0019003 | GDP binding | 1.40e-01 | 1.00e+00 | 2.737 | 1 | 1 | 48 |
| GO:0005070 | SH3/SH2 adaptor activity | 1.42e-01 | 1.00e+00 | 2.707 | 1 | 1 | 49 |
| GO:0045111 | intermediate filament cytoskeleton | 1.42e-01 | 1.00e+00 | 2.707 | 1 | 1 | 49 |
| GO:0001948 | glycoprotein binding | 1.45e-01 | 1.00e+00 | 2.678 | 1 | 2 | 50 |
| GO:0035690 | cellular response to drug | 1.45e-01 | 1.00e+00 | 2.678 | 1 | 3 | 50 |
| GO:0007030 | Golgi organization | 1.45e-01 | 1.00e+00 | 2.678 | 1 | 3 | 50 |
| GO:0005765 | lysosomal membrane | 1.48e-01 | 1.00e+00 | 1.554 | 2 | 2 | 218 |
| GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1.48e-01 | 1.00e+00 | 2.650 | 1 | 1 | 51 |
| GO:0000902 | cell morphogenesis | 1.48e-01 | 1.00e+00 | 2.650 | 1 | 1 | 51 |
| GO:0030900 | forebrain development | 1.48e-01 | 1.00e+00 | 2.650 | 1 | 1 | 51 |
| GO:0008202 | steroid metabolic process | 1.50e-01 | 1.00e+00 | 2.622 | 1 | 1 | 52 |
| GO:0006184 | GTP catabolic process | 1.51e-01 | 1.00e+00 | 1.541 | 2 | 6 | 220 |
| GO:0006952 | defense response | 1.53e-01 | 1.00e+00 | 2.594 | 1 | 1 | 53 |
| GO:0030175 | filopodium | 1.53e-01 | 1.00e+00 | 2.594 | 1 | 2 | 53 |
| GO:0003725 | double-stranded RNA binding | 1.56e-01 | 1.00e+00 | 2.567 | 1 | 2 | 54 |
| GO:0009612 | response to mechanical stimulus | 1.56e-01 | 1.00e+00 | 2.567 | 1 | 1 | 54 |
| GO:0000186 | activation of MAPKK activity | 1.56e-01 | 1.00e+00 | 2.567 | 1 | 1 | 54 |
| GO:0050679 | positive regulation of epithelial cell proliferation | 1.56e-01 | 1.00e+00 | 2.567 | 1 | 2 | 54 |
| GO:0097193 | intrinsic apoptotic signaling pathway | 1.58e-01 | 1.00e+00 | 2.541 | 1 | 2 | 55 |
| GO:0002039 | p53 binding | 1.58e-01 | 1.00e+00 | 2.541 | 1 | 1 | 55 |
| GO:0046330 | positive regulation of JNK cascade | 1.58e-01 | 1.00e+00 | 2.541 | 1 | 1 | 55 |
| GO:0019221 | cytokine-mediated signaling pathway | 1.61e-01 | 1.00e+00 | 1.477 | 2 | 5 | 230 |
| GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment | 1.64e-01 | 1.00e+00 | 2.489 | 1 | 1 | 57 |
| GO:0006879 | cellular iron ion homeostasis | 1.64e-01 | 1.00e+00 | 2.489 | 1 | 2 | 57 |
| GO:0005643 | nuclear pore | 1.69e-01 | 1.00e+00 | 2.439 | 1 | 1 | 59 |
| GO:0010976 | positive regulation of neuron projection development | 1.71e-01 | 1.00e+00 | 2.415 | 1 | 2 | 60 |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1.74e-01 | 1.00e+00 | 2.391 | 1 | 1 | 61 |
| GO:0007596 | blood coagulation | 1.76e-01 | 1.00e+00 | 1.049 | 3 | 5 | 464 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 1.77e-01 | 1.00e+00 | 2.368 | 1 | 1 | 62 |
| GO:0000151 | ubiquitin ligase complex | 1.79e-01 | 1.00e+00 | 2.345 | 1 | 1 | 63 |
| GO:0005901 | caveola | 1.79e-01 | 1.00e+00 | 2.345 | 1 | 1 | 63 |
| GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1.79e-01 | 1.00e+00 | 2.345 | 1 | 2 | 63 |
| GO:0032869 | cellular response to insulin stimulus | 1.82e-01 | 1.00e+00 | 2.322 | 1 | 2 | 64 |
| GO:0016491 | oxidoreductase activity | 1.84e-01 | 1.00e+00 | 2.300 | 1 | 1 | 65 |
| GO:0043025 | neuronal cell body | 1.88e-01 | 1.00e+00 | 1.333 | 2 | 5 | 254 |
| GO:0030141 | secretory granule | 1.90e-01 | 1.00e+00 | 2.256 | 1 | 2 | 67 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.90e-01 | 1.00e+00 | 1.322 | 2 | 3 | 256 |
| GO:0006665 | sphingolipid metabolic process | 1.92e-01 | 1.00e+00 | 2.235 | 1 | 1 | 68 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 1.95e-01 | 1.00e+00 | 0.979 | 3 | 8 | 487 |
| GO:0035264 | multicellular organism growth | 1.97e-01 | 1.00e+00 | 2.193 | 1 | 1 | 70 |
| GO:0042393 | histone binding | 2.00e-01 | 1.00e+00 | 2.172 | 1 | 1 | 71 |
| GO:0006281 | DNA repair | 2.00e-01 | 1.00e+00 | 1.278 | 2 | 5 | 264 |
| GO:0001503 | ossification | 2.00e-01 | 1.00e+00 | 2.172 | 1 | 2 | 71 |
| GO:0000165 | MAPK cascade | 2.02e-01 | 1.00e+00 | 2.152 | 1 | 2 | 72 |
| GO:0000785 | chromatin | 2.05e-01 | 1.00e+00 | 2.132 | 1 | 2 | 73 |
| GO:0055086 | nucleobase-containing small molecule metabolic process | 2.05e-01 | 1.00e+00 | 2.132 | 1 | 2 | 73 |
| GO:0042826 | histone deacetylase binding | 2.07e-01 | 1.00e+00 | 2.113 | 1 | 1 | 74 |
| GO:0000166 | nucleotide binding | 2.09e-01 | 1.00e+00 | 1.235 | 2 | 2 | 272 |
| GO:0051897 | positive regulation of protein kinase B signaling | 2.10e-01 | 1.00e+00 | 2.093 | 1 | 1 | 75 |
| GO:0060070 | canonical Wnt signaling pathway | 2.10e-01 | 1.00e+00 | 2.093 | 1 | 2 | 75 |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 2.10e-01 | 1.00e+00 | 2.093 | 1 | 1 | 75 |
| GO:0005975 | carbohydrate metabolic process | 2.11e-01 | 1.00e+00 | 1.224 | 2 | 3 | 274 |
| GO:0044325 | ion channel binding | 2.15e-01 | 1.00e+00 | 2.055 | 1 | 3 | 77 |
| GO:0055085 | transmembrane transport | 2.16e-01 | 1.00e+00 | 0.901 | 3 | 3 | 514 |
| GO:0071013 | catalytic step 2 spliceosome | 2.20e-01 | 1.00e+00 | 2.018 | 1 | 1 | 79 |
| GO:0031902 | late endosome membrane | 2.22e-01 | 1.00e+00 | 2.000 | 1 | 1 | 80 |
| GO:0048471 | perinuclear region of cytoplasm | 2.24e-01 | 1.00e+00 | 0.876 | 3 | 9 | 523 |
| GO:0045177 | apical part of cell | 2.27e-01 | 1.00e+00 | 1.964 | 1 | 1 | 82 |
| GO:0001726 | ruffle | 2.27e-01 | 1.00e+00 | 1.964 | 1 | 1 | 82 |
| GO:0019899 | enzyme binding | 2.27e-01 | 1.00e+00 | 1.152 | 2 | 5 | 288 |
| GO:0004713 | protein tyrosine kinase activity | 2.27e-01 | 1.00e+00 | 1.964 | 1 | 1 | 82 |
| GO:0030336 | negative regulation of cell migration | 2.29e-01 | 1.00e+00 | 1.947 | 1 | 2 | 83 |
| GO:0007264 | small GTPase mediated signal transduction | 2.29e-01 | 1.00e+00 | 1.142 | 2 | 7 | 290 |
| GO:0005929 | cilium | 2.32e-01 | 1.00e+00 | 1.930 | 1 | 2 | 84 |
| GO:0005179 | hormone activity | 2.32e-01 | 1.00e+00 | 1.930 | 1 | 1 | 84 |
| GO:0043234 | protein complex | 2.41e-01 | 1.00e+00 | 1.093 | 2 | 9 | 300 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2.41e-01 | 1.00e+00 | 1.863 | 1 | 3 | 88 |
| GO:0042593 | glucose homeostasis | 2.44e-01 | 1.00e+00 | 1.846 | 1 | 1 | 89 |
| GO:0042384 | cilium assembly | 2.46e-01 | 1.00e+00 | 1.830 | 1 | 2 | 90 |
| GO:0005777 | peroxisome | 2.46e-01 | 1.00e+00 | 1.830 | 1 | 1 | 90 |
| GO:0005524 | ATP binding | 2.46e-01 | 1.00e+00 | 0.504 | 6 | 19 | 1354 |
| GO:0003690 | double-stranded DNA binding | 2.48e-01 | 1.00e+00 | 1.814 | 1 | 2 | 91 |
| GO:0016337 | single organismal cell-cell adhesion | 2.51e-01 | 1.00e+00 | 1.798 | 1 | 2 | 92 |
| GO:0042470 | melanosome | 2.51e-01 | 1.00e+00 | 1.798 | 1 | 2 | 92 |
| GO:0005200 | structural constituent of cytoskeleton | 2.53e-01 | 1.00e+00 | 1.783 | 1 | 6 | 93 |
| GO:0004674 | protein serine/threonine kinase activity | 2.55e-01 | 1.00e+00 | 1.037 | 2 | 6 | 312 |
| GO:0005770 | late endosome | 2.55e-01 | 1.00e+00 | 1.767 | 1 | 1 | 94 |
| GO:0035556 | intracellular signal transduction | 2.61e-01 | 1.00e+00 | 1.014 | 2 | 5 | 317 |
| GO:0006355 | regulation of transcription, DNA-templated | 2.61e-01 | 1.00e+00 | 0.535 | 5 | 10 | 1104 |
| GO:0005178 | integrin binding | 2.62e-01 | 1.00e+00 | 1.722 | 1 | 1 | 97 |
| GO:0007411 | axon guidance | 2.72e-01 | 1.00e+00 | 0.969 | 2 | 3 | 327 |
| GO:0005525 | GTP binding | 2.73e-01 | 1.00e+00 | 0.964 | 2 | 6 | 328 |
| GO:0008283 | cell proliferation | 2.77e-01 | 1.00e+00 | 0.951 | 2 | 4 | 331 |
| GO:0014069 | postsynaptic density | 2.83e-01 | 1.00e+00 | 1.594 | 1 | 3 | 106 |
| GO:0006935 | chemotaxis | 2.88e-01 | 1.00e+00 | 1.567 | 1 | 1 | 108 |
| GO:0030496 | midbody | 2.90e-01 | 1.00e+00 | 1.554 | 1 | 4 | 109 |
| GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 2.92e-01 | 1.00e+00 | 1.541 | 1 | 1 | 110 |
| GO:0050900 | leukocyte migration | 2.94e-01 | 1.00e+00 | 1.528 | 1 | 1 | 111 |
| GO:0020037 | heme binding | 2.94e-01 | 1.00e+00 | 1.528 | 1 | 1 | 111 |
| GO:0015630 | microtubule cytoskeleton | 2.97e-01 | 1.00e+00 | 1.515 | 1 | 4 | 112 |
| GO:0032496 | response to lipopolysaccharide | 3.18e-01 | 1.00e+00 | 1.391 | 1 | 1 | 122 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 3.21e-01 | 1.00e+00 | 1.380 | 1 | 3 | 123 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 3.29e-01 | 1.00e+00 | 1.333 | 1 | 3 | 127 |
| GO:0005794 | Golgi apparatus | 3.32e-01 | 1.00e+00 | 0.563 | 3 | 8 | 650 |
| GO:0030036 | actin cytoskeleton organization | 3.33e-01 | 1.00e+00 | 1.311 | 1 | 3 | 129 |
| GO:0030027 | lamellipodium | 3.35e-01 | 1.00e+00 | 1.300 | 1 | 3 | 130 |
| GO:0046983 | protein dimerization activity | 3.37e-01 | 1.00e+00 | 1.289 | 1 | 3 | 131 |
| GO:0018108 | peptidyl-tyrosine phosphorylation | 3.42e-01 | 1.00e+00 | 1.267 | 1 | 1 | 133 |
| GO:0007165 | signal transduction | 3.45e-01 | 1.00e+00 | 0.430 | 4 | 7 | 950 |
| GO:0016055 | Wnt signaling pathway | 3.58e-01 | 1.00e+00 | 1.182 | 1 | 3 | 141 |
| GO:0005911 | cell-cell junction | 3.60e-01 | 1.00e+00 | 1.172 | 1 | 2 | 142 |
| GO:0000139 | Golgi membrane | 3.66e-01 | 1.00e+00 | 0.650 | 2 | 7 | 408 |
| GO:0007166 | cell surface receptor signaling pathway | 3.74e-01 | 1.00e+00 | 1.103 | 1 | 1 | 149 |
| GO:0044281 | small molecule metabolic process | 3.81e-01 | 1.00e+00 | 0.305 | 5 | 16 | 1295 |
| GO:0006366 | transcription from RNA polymerase II promoter | 3.85e-01 | 1.00e+00 | 0.591 | 2 | 3 | 425 |
| GO:0046777 | protein autophosphorylation | 3.92e-01 | 1.00e+00 | 1.018 | 1 | 1 | 158 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 4.11e-01 | 1.00e+00 | 0.930 | 1 | 4 | 168 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 4.16e-01 | 1.00e+00 | 0.360 | 3 | 9 | 748 |
| GO:0007420 | brain development | 4.18e-01 | 1.00e+00 | 0.896 | 1 | 3 | 172 |
| GO:0016607 | nuclear speck | 4.23e-01 | 1.00e+00 | 0.871 | 1 | 2 | 175 |
| GO:0031965 | nuclear membrane | 4.25e-01 | 1.00e+00 | 0.863 | 1 | 2 | 176 |
| GO:0007049 | cell cycle | 4.27e-01 | 1.00e+00 | 0.854 | 1 | 4 | 177 |
| GO:0006468 | protein phosphorylation | 4.31e-01 | 1.00e+00 | 0.455 | 2 | 6 | 467 |
| GO:0031625 | ubiquitin protein ligase binding | 4.32e-01 | 1.00e+00 | 0.830 | 1 | 4 | 180 |
| GO:0005764 | lysosome | 4.36e-01 | 1.00e+00 | 0.814 | 1 | 2 | 182 |
| GO:0042802 | identical protein binding | 4.57e-01 | 1.00e+00 | 0.382 | 2 | 4 | 491 |
| GO:0001525 | angiogenesis | 4.67e-01 | 1.00e+00 | 0.678 | 1 | 3 | 200 |
| GO:0001701 | in utero embryonic development | 4.84e-01 | 1.00e+00 | 0.608 | 1 | 2 | 210 |
| GO:0004871 | signal transducer activity | 4.94e-01 | 1.00e+00 | 0.567 | 1 | 1 | 216 |
| GO:0016874 | ligase activity | 5.06e-01 | 1.00e+00 | 0.515 | 1 | 2 | 224 |
| GO:0008380 | RNA splicing | 5.19e-01 | 1.00e+00 | 0.464 | 1 | 5 | 232 |
| GO:0030425 | dendrite | 5.31e-01 | 1.00e+00 | 0.415 | 1 | 3 | 240 |
| GO:0005102 | receptor binding | 5.71e-01 | 1.00e+00 | 0.256 | 1 | 2 | 268 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 5.80e-01 | 1.00e+00 | 0.219 | 1 | 2 | 275 |
| GO:0007283 | spermatogenesis | 5.82e-01 | 1.00e+00 | 0.213 | 1 | 2 | 276 |
| GO:0042493 | response to drug | 5.97e-01 | 1.00e+00 | 0.152 | 1 | 2 | 288 |
| GO:0016567 | protein ubiquitination | 6.11e-01 | 1.00e+00 | 0.098 | 1 | 3 | 299 |
| GO:0005743 | mitochondrial inner membrane | 6.12e-01 | 1.00e+00 | 0.093 | 1 | 1 | 300 |
| GO:0006955 | immune response | 6.18e-01 | 1.00e+00 | 0.069 | 1 | 2 | 305 |
| GO:0003677 | DNA binding | 6.19e-01 | 1.00e+00 | -0.078 | 4 | 14 | 1351 |
| GO:0005856 | cytoskeleton | 6.26e-01 | 1.00e+00 | 0.041 | 1 | 6 | 311 |
| GO:0005575 | cellular_component | 6.39e-01 | 1.00e+00 | -0.009 | 1 | 2 | 322 |
| GO:0030154 | cell differentiation | 6.42e-01 | 1.00e+00 | -0.022 | 1 | 3 | 325 |
| GO:0007275 | multicellular organismal development | 6.63e-01 | 1.00e+00 | -0.104 | 1 | 2 | 344 |
| GO:0015031 | protein transport | 6.77e-01 | 1.00e+00 | -0.158 | 1 | 4 | 357 |
| GO:0005886 | plasma membrane | 6.81e-01 | 1.00e+00 | -0.147 | 8 | 24 | 2834 |
| GO:0046872 | metal ion binding | 6.83e-01 | 1.00e+00 | -0.195 | 4 | 14 | 1465 |
| GO:0008285 | negative regulation of cell proliferation | 6.87e-01 | 1.00e+00 | -0.198 | 1 | 3 | 367 |
| GO:0007155 | cell adhesion | 7.04e-01 | 1.00e+00 | -0.263 | 1 | 3 | 384 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 7.39e-01 | 1.00e+00 | -0.406 | 1 | 2 | 424 |
| GO:0008150 | biological_process | 7.67e-01 | 1.00e+00 | -0.520 | 1 | 3 | 459 |
| GO:0055114 | oxidation-reduction process | 7.83e-01 | 1.00e+00 | -0.588 | 1 | 2 | 481 |
| GO:0008270 | zinc ion binding | 8.55e-01 | 1.00e+00 | -0.737 | 2 | 7 | 1067 |
| GO:0005783 | endoplasmic reticulum | 8.57e-01 | 1.00e+00 | -0.931 | 1 | 6 | 610 |
| GO:0042803 | protein homodimerization activity | 8.60e-01 | 1.00e+00 | -0.947 | 1 | 4 | 617 |
| GO:0005615 | extracellular space | 9.62e-01 | 1.00e+00 | -1.658 | 1 | 3 | 1010 |