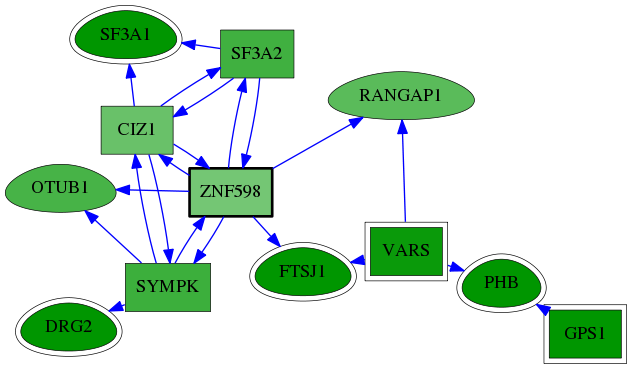
| GO:0000389 | mRNA 3'-splice site recognition | 8.30e-06 | 8.10e-02 | 8.668 | 2 | 2 | 4 |
| GO:0004832 | valine-tRNA ligase activity | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 1 |
| GO:0002128 | tRNA nucleoside ribose methylation | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 1 |
| GO:0006438 | valyl-tRNA aminoacylation | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 1 |
| GO:0005098 | Ran GTPase activator activity | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 1 |
| GO:0008175 | tRNA methyltransferase activity | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 1 |
| GO:0005681 | spliceosomal complex | 1.98e-03 | 1.00e+00 | 4.887 | 2 | 8 | 55 |
| GO:0071013 | catalytic step 2 spliceosome | 2.43e-03 | 1.00e+00 | 4.738 | 2 | 5 | 61 |
| GO:0019784 | NEDD8-specific protease activity | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 2 |
| GO:0005684 | U2-type spliceosomal complex | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 2 | 2 |
| GO:1901315 | negative regulation of histone H2A K63-linked ubiquitination | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 2 | 3 |
| GO:0032853 | positive regulation of Ran GTPase activity | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 3 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 5 |
| GO:0002161 | aminoacyl-tRNA editing activity | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 5 |
| GO:2000780 | negative regulation of double-strand break repair | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 2 | 5 |
| GO:0050847 | progesterone receptor signaling pathway | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 2 | 6 |
| GO:0002181 | cytoplasmic translation | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 6 |
| GO:0046826 | negative regulation of protein export from nucleus | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 6 |
| GO:0006397 | mRNA processing | 7.71e-03 | 1.00e+00 | 3.887 | 2 | 12 | 110 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 1 | 8 |
| GO:0010388 | cullin deneddylation | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 4 | 8 |
| GO:0008242 | omega peptidase activity | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 1 | 8 |
| GO:0006450 | regulation of translational fidelity | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 1 | 8 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.03e-02 | 1.00e+00 | 3.668 | 2 | 15 | 128 |
| GO:0071354 | cellular response to interleukin-6 | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 1 | 9 |
| GO:0005095 | GTPase inhibitor activity | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 1 | 10 |
| GO:0035307 | positive regulation of protein dephosphorylation | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 1 | 10 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 4 | 11 |
| GO:0071108 | protein K48-linked deubiquitination | 1.59e-02 | 1.00e+00 | 5.968 | 1 | 1 | 13 |
| GO:0008380 | RNA splicing | 1.68e-02 | 1.00e+00 | 3.302 | 2 | 21 | 165 |
| GO:0016575 | histone deacetylation | 1.71e-02 | 1.00e+00 | 5.861 | 1 | 2 | 14 |
| GO:0005654 | nucleoplasm | 1.77e-02 | 1.00e+00 | 1.894 | 4 | 68 | 876 |
| GO:0000188 | inactivation of MAPK activity | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 1 | 15 |
| GO:0030532 | small nuclear ribonucleoprotein complex | 2.07e-02 | 1.00e+00 | 5.581 | 1 | 1 | 17 |
| GO:0005737 | cytoplasm | 2.16e-02 | 1.00e+00 | 1.113 | 7 | 127 | 2633 |
| GO:0006378 | mRNA polyadenylation | 2.19e-02 | 1.00e+00 | 5.499 | 1 | 3 | 18 |
| GO:0071897 | DNA biosynthetic process | 2.43e-02 | 1.00e+00 | 5.347 | 1 | 2 | 20 |
| GO:0010467 | gene expression | 2.48e-02 | 1.00e+00 | 2.190 | 3 | 49 | 535 |
| GO:0002376 | immune system process | 2.79e-02 | 1.00e+00 | 5.145 | 1 | 2 | 23 |
| GO:0008180 | COP9 signalosome | 2.91e-02 | 1.00e+00 | 5.083 | 1 | 6 | 24 |
| GO:0006418 | tRNA aminoacylation for protein translation | 3.51e-02 | 1.00e+00 | 4.810 | 1 | 4 | 29 |
| GO:0007165 | signal transduction | 3.61e-02 | 1.00e+00 | 1.982 | 3 | 24 | 618 |
| GO:0043130 | ubiquitin binding | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 3 | 30 |
| GO:0007254 | JNK cascade | 4.10e-02 | 1.00e+00 | 4.581 | 1 | 1 | 34 |
| GO:0005739 | mitochondrion | 4.26e-02 | 1.00e+00 | 1.889 | 3 | 32 | 659 |
| GO:0008270 | zinc ion binding | 5.45e-02 | 1.00e+00 | 1.748 | 3 | 39 | 727 |
| GO:0004843 | ubiquitin-specific protease activity | 5.51e-02 | 1.00e+00 | 4.145 | 1 | 2 | 46 |
| GO:0005643 | nuclear pore | 6.09e-02 | 1.00e+00 | 3.996 | 1 | 12 | 51 |
| GO:0000776 | kinetochore | 6.56e-02 | 1.00e+00 | 3.887 | 1 | 8 | 55 |
| GO:0000777 | condensed chromosome kinetochore | 6.79e-02 | 1.00e+00 | 3.836 | 1 | 6 | 57 |
| GO:0044822 | poly(A) RNA binding | 6.88e-02 | 1.00e+00 | 1.611 | 3 | 45 | 799 |
| GO:0042826 | histone deacetylase binding | 6.90e-02 | 1.00e+00 | 3.810 | 1 | 5 | 58 |
| GO:0005923 | tight junction | 7.59e-02 | 1.00e+00 | 3.668 | 1 | 2 | 64 |
| GO:0003676 | nucleic acid binding | 7.82e-02 | 1.00e+00 | 3.624 | 1 | 4 | 66 |
| GO:0005515 | protein binding | 7.83e-02 | 1.00e+00 | 0.659 | 8 | 198 | 4124 |
| GO:0000922 | spindle pole | 8.05e-02 | 1.00e+00 | 3.581 | 1 | 4 | 68 |
| GO:0030308 | negative regulation of cell growth | 8.28e-02 | 1.00e+00 | 3.539 | 1 | 3 | 70 |
| GO:0001649 | osteoblast differentiation | 8.73e-02 | 1.00e+00 | 3.459 | 1 | 5 | 74 |
| GO:0042981 | regulation of apoptotic process | 1.29e-01 | 1.00e+00 | 2.861 | 1 | 4 | 112 |
| GO:0007049 | cell cycle | 1.33e-01 | 1.00e+00 | 2.823 | 1 | 8 | 115 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.39e-01 | 1.00e+00 | 2.750 | 1 | 11 | 121 |
| GO:0031965 | nuclear membrane | 1.49e-01 | 1.00e+00 | 2.646 | 1 | 8 | 130 |
| GO:0031625 | ubiquitin protein ligase binding | 1.57e-01 | 1.00e+00 | 2.560 | 1 | 7 | 138 |
| GO:0044212 | transcription regulatory region DNA binding | 1.62e-01 | 1.00e+00 | 2.509 | 1 | 13 | 143 |
| GO:0005856 | cytoskeleton | 2.10e-01 | 1.00e+00 | 2.099 | 1 | 8 | 190 |
| GO:0005743 | mitochondrial inner membrane | 2.17e-01 | 1.00e+00 | 2.046 | 1 | 9 | 197 |
| GO:0005525 | GTP binding | 2.17e-01 | 1.00e+00 | 2.046 | 1 | 12 | 197 |
| GO:0006281 | DNA repair | 2.23e-01 | 1.00e+00 | 2.003 | 1 | 24 | 203 |
| GO:0019899 | enzyme binding | 2.30e-01 | 1.00e+00 | 1.954 | 1 | 7 | 210 |
| GO:0007155 | cell adhesion | 2.62e-01 | 1.00e+00 | 1.738 | 1 | 8 | 244 |
| GO:0003723 | RNA binding | 2.65e-01 | 1.00e+00 | 1.720 | 1 | 22 | 247 |
| GO:0006508 | proteolysis | 2.80e-01 | 1.00e+00 | 1.624 | 1 | 11 | 264 |
| GO:0008285 | negative regulation of cell proliferation | 2.92e-01 | 1.00e+00 | 1.555 | 1 | 11 | 277 |
| GO:0000278 | mitotic cell cycle | 3.25e-01 | 1.00e+00 | 1.374 | 1 | 28 | 314 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.27e-01 | 1.00e+00 | 1.360 | 1 | 17 | 317 |
| GO:0048471 | perinuclear region of cytoplasm | 3.50e-01 | 1.00e+00 | 1.242 | 1 | 11 | 344 |
| GO:0005634 | nucleus | 3.66e-01 | 1.00e+00 | 0.326 | 5 | 158 | 3246 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 3.77e-01 | 1.00e+00 | 1.110 | 1 | 21 | 377 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 4.37e-01 | 1.00e+00 | 0.836 | 1 | 29 | 456 |
| GO:0016020 | membrane | 4.47e-01 | 1.00e+00 | 0.431 | 2 | 56 | 1207 |
| GO:0005730 | nucleolus | 4.52e-01 | 1.00e+00 | 0.419 | 2 | 74 | 1217 |
| GO:0005887 | integral component of plasma membrane | 5.17e-01 | 1.00e+00 | 0.501 | 1 | 8 | 575 |
| GO:0006355 | regulation of transcription, DNA-templated | 5.99e-01 | 1.00e+00 | 0.187 | 1 | 31 | 715 |
| GO:0070062 | extracellular vesicular exosome | 6.24e-01 | 1.00e+00 | -0.012 | 2 | 51 | 1641 |
| GO:0005829 | cytosol | 6.74e-01 | 1.00e+00 | -0.135 | 2 | 88 | 1787 |
| GO:0005524 | ATP binding | 6.83e-01 | 1.00e+00 | -0.132 | 1 | 37 | 892 |
| GO:0005886 | plasma membrane | 9.11e-01 | 1.00e+00 | -1.132 | 1 | 46 | 1784 |