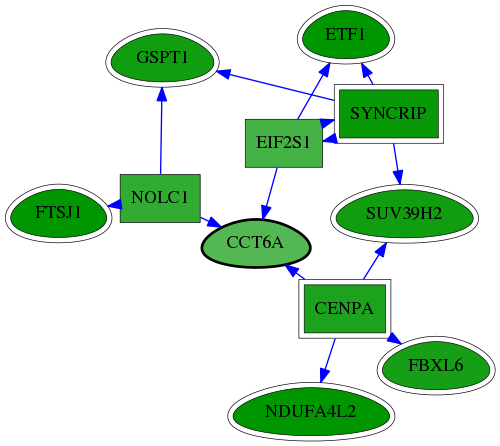
| GO:0003747 | translation release factor activity | 1.15e-06 | 1.13e-02 | 9.794 | 2 | 2 | 2 |
| GO:0006479 | protein methylation | 6.31e-05 | 6.16e-01 | 7.335 | 2 | 2 | 11 |
| GO:0044822 | poly(A) RNA binding | 9.51e-05 | 9.28e-01 | 2.737 | 6 | 45 | 799 |
| GO:0043022 | ribosome binding | 1.55e-04 | 1.00e+00 | 6.707 | 2 | 2 | 17 |
| GO:0000775 | chromosome, centromeric region | 9.24e-04 | 1.00e+00 | 5.436 | 2 | 6 | 41 |
| GO:0000939 | condensed chromosome inner kinetochore | 1.13e-03 | 1.00e+00 | 9.794 | 1 | 1 | 1 |
| GO:0016149 | translation release factor activity, codon specific | 1.13e-03 | 1.00e+00 | 9.794 | 1 | 1 | 1 |
| GO:0008079 | translation termination factor activity | 1.13e-03 | 1.00e+00 | 9.794 | 1 | 1 | 1 |
| GO:0002128 | tRNA nucleoside ribose methylation | 1.13e-03 | 1.00e+00 | 9.794 | 1 | 1 | 1 |
| GO:0008175 | tRNA methyltransferase activity | 1.13e-03 | 1.00e+00 | 9.794 | 1 | 1 | 1 |
| GO:0006415 | translational termination | 2.17e-03 | 1.00e+00 | 4.817 | 2 | 2 | 63 |
| GO:0043558 | regulation of translational initiation in response to stress | 2.25e-03 | 1.00e+00 | 8.794 | 1 | 1 | 2 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 2 | 3 |
| GO:0036123 | histone H3-K9 dimethylation | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 3 |
| GO:0000778 | condensed nuclear chromosome kinetochore | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 2 | 3 |
| GO:0071459 | protein localization to chromosome, centromeric region | 3.38e-03 | 1.00e+00 | 8.209 | 1 | 1 | 3 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 3.82e-03 | 1.00e+00 | 4.402 | 2 | 5 | 84 |
| GO:0006449 | regulation of translational termination | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 4 |
| GO:0007000 | nucleolus organization | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 4 |
| GO:0006333 | chromatin assembly or disassembly | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 1 | 4 |
| GO:0036124 | histone H3-K9 trimethylation | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 4 |
| GO:0097452 | GAIT complex | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 4 |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | 4.50e-03 | 1.00e+00 | 7.794 | 1 | 2 | 4 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 2 | 5 |
| GO:0044267 | cellular protein metabolic process | 6.18e-03 | 1.00e+00 | 2.920 | 3 | 14 | 352 |
| GO:0002181 | cytoplasmic translation | 6.74e-03 | 1.00e+00 | 7.209 | 1 | 1 | 6 |
| GO:0070937 | CRD-mediated mRNA stability complex | 6.74e-03 | 1.00e+00 | 7.209 | 1 | 2 | 6 |
| GO:0042754 | negative regulation of circadian rhythm | 6.74e-03 | 1.00e+00 | 7.209 | 1 | 2 | 6 |
| GO:0046974 | histone methyltransferase activity (H3-K9 specific) | 6.74e-03 | 1.00e+00 | 7.209 | 1 | 1 | 6 |
| GO:0002199 | zona pellucida receptor complex | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 4 | 7 |
| GO:0007140 | male meiosis | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 7 |
| GO:0000780 | condensed nuclear chromosome, centromeric region | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 1 | 7 |
| GO:0005832 | chaperonin-containing T-complex | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 4 | 7 |
| GO:0051382 | kinetochore assembly | 7.86e-03 | 1.00e+00 | 6.987 | 1 | 2 | 7 |
| GO:0008143 | poly(A) binding | 1.12e-02 | 1.00e+00 | 6.472 | 1 | 2 | 10 |
| GO:0000132 | establishment of mitotic spindle orientation | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 2 | 12 |
| GO:0006412 | translation | 1.63e-02 | 1.00e+00 | 3.318 | 2 | 12 | 178 |
| GO:0007339 | binding of sperm to zona pellucida | 1.68e-02 | 1.00e+00 | 5.887 | 1 | 4 | 15 |
| GO:0005525 | GTP binding | 1.98e-02 | 1.00e+00 | 3.172 | 2 | 12 | 197 |
| GO:0005844 | polysome | 2.12e-02 | 1.00e+00 | 5.546 | 1 | 2 | 19 |
| GO:0071346 | cellular response to interferon-gamma | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 2 | 20 |
| GO:0005720 | nuclear heterochromatin | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 3 | 20 |
| GO:0034080 | CENP-A containing nucleosome assembly | 2.23e-02 | 1.00e+00 | 5.472 | 1 | 2 | 20 |
| GO:0051084 | 'de novo' posttranslational protein folding | 2.45e-02 | 1.00e+00 | 5.335 | 1 | 4 | 22 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 2.78e-02 | 1.00e+00 | 5.150 | 1 | 3 | 25 |
| GO:0010494 | cytoplasmic stress granule | 2.89e-02 | 1.00e+00 | 5.094 | 1 | 4 | 26 |
| GO:0003723 | RNA binding | 3.02e-02 | 1.00e+00 | 2.846 | 2 | 22 | 247 |
| GO:0001669 | acrosomal vesicle | 3.11e-02 | 1.00e+00 | 4.987 | 1 | 1 | 28 |
| GO:0003682 | chromatin binding | 3.13e-02 | 1.00e+00 | 2.817 | 2 | 15 | 252 |
| GO:0015030 | Cajal body | 3.55e-02 | 1.00e+00 | 4.794 | 1 | 3 | 32 |
| GO:0017148 | negative regulation of translation | 3.66e-02 | 1.00e+00 | 4.750 | 1 | 5 | 33 |
| GO:0000786 | nucleosome | 3.77e-02 | 1.00e+00 | 4.707 | 1 | 2 | 34 |
| GO:0044297 | cell body | 3.87e-02 | 1.00e+00 | 4.665 | 1 | 5 | 35 |
| GO:0003743 | translation initiation factor activity | 4.09e-02 | 1.00e+00 | 4.585 | 1 | 8 | 37 |
| GO:0005515 | protein binding | 4.12e-02 | 1.00e+00 | 0.784 | 8 | 198 | 4124 |
| GO:0048511 | rhythmic process | 4.20e-02 | 1.00e+00 | 4.546 | 1 | 5 | 38 |
| GO:0006396 | RNA processing | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 4 | 40 |
| GO:0006987 | activation of signaling protein activity involved in unfolded protein response | 5.17e-02 | 1.00e+00 | 4.239 | 1 | 1 | 47 |
| GO:0006334 | nucleosome assembly | 5.60e-02 | 1.00e+00 | 4.122 | 1 | 4 | 51 |
| GO:0006338 | chromatin remodeling | 6.03e-02 | 1.00e+00 | 4.013 | 1 | 7 | 55 |
| GO:0000785 | chromatin | 6.45e-02 | 1.00e+00 | 3.911 | 1 | 6 | 59 |
| GO:0030968 | endoplasmic reticulum unfolded protein response | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 1 | 61 |
| GO:0071013 | catalytic step 2 spliceosome | 6.66e-02 | 1.00e+00 | 3.863 | 1 | 5 | 61 |
| GO:0006364 | rRNA processing | 7.30e-02 | 1.00e+00 | 3.728 | 1 | 2 | 67 |
| GO:0051082 | unfolded protein binding | 7.61e-02 | 1.00e+00 | 3.665 | 1 | 5 | 70 |
| GO:0001649 | osteoblast differentiation | 8.03e-02 | 1.00e+00 | 3.585 | 1 | 5 | 74 |
| GO:0016032 | viral process | 8.12e-02 | 1.00e+00 | 2.053 | 2 | 30 | 428 |
| GO:0071456 | cellular response to hypoxia | 8.24e-02 | 1.00e+00 | 3.546 | 1 | 2 | 76 |
| GO:0030529 | ribonucleoprotein complex | 8.86e-02 | 1.00e+00 | 3.436 | 1 | 5 | 82 |
| GO:0006457 | protein folding | 1.04e-01 | 1.00e+00 | 3.194 | 1 | 5 | 97 |
| GO:0006413 | translational initiation | 1.06e-01 | 1.00e+00 | 3.165 | 1 | 6 | 99 |
| GO:0046777 | protein autophosphorylation | 1.13e-01 | 1.00e+00 | 3.066 | 1 | 3 | 106 |
| GO:0010467 | gene expression | 1.19e-01 | 1.00e+00 | 1.731 | 2 | 49 | 535 |
| GO:0007049 | cell cycle | 1.22e-01 | 1.00e+00 | 2.948 | 1 | 8 | 115 |
| GO:0005829 | cytosol | 1.25e-01 | 1.00e+00 | 0.991 | 4 | 88 | 1787 |
| GO:0003924 | GTPase activity | 1.26e-01 | 1.00e+00 | 2.899 | 1 | 7 | 119 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.32e-01 | 1.00e+00 | 2.828 | 1 | 4 | 125 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.35e-01 | 1.00e+00 | 2.794 | 1 | 15 | 128 |
| GO:0006184 | GTP catabolic process | 1.37e-01 | 1.00e+00 | 2.772 | 1 | 7 | 130 |
| GO:0005622 | intracellular | 1.52e-01 | 1.00e+00 | 2.614 | 1 | 2 | 145 |
| GO:0005874 | microtubule | 1.69e-01 | 1.00e+00 | 2.445 | 1 | 10 | 163 |
| GO:0007067 | mitotic nuclear division | 1.71e-01 | 1.00e+00 | 2.428 | 1 | 9 | 165 |
| GO:0008380 | RNA splicing | 1.71e-01 | 1.00e+00 | 2.428 | 1 | 21 | 165 |
| GO:0016071 | mRNA metabolic process | 1.76e-01 | 1.00e+00 | 2.385 | 1 | 10 | 170 |
| GO:0000166 | nucleotide binding | 1.81e-01 | 1.00e+00 | 2.335 | 1 | 13 | 176 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.84e-01 | 1.00e+00 | 2.310 | 1 | 17 | 179 |
| GO:0016070 | RNA metabolic process | 1.93e-01 | 1.00e+00 | 2.239 | 1 | 10 | 188 |
| GO:0016567 | protein ubiquitination | 1.93e-01 | 1.00e+00 | 2.239 | 1 | 14 | 188 |
| GO:0030154 | cell differentiation | 2.07e-01 | 1.00e+00 | 2.122 | 1 | 5 | 204 |
| GO:0005654 | nucleoplasm | 2.59e-01 | 1.00e+00 | 1.019 | 2 | 68 | 876 |
| GO:0006508 | proteolysis | 2.60e-01 | 1.00e+00 | 1.750 | 1 | 11 | 264 |
| GO:0046982 | protein heterodimerization activity | 2.61e-01 | 1.00e+00 | 1.744 | 1 | 11 | 265 |
| GO:0005524 | ATP binding | 2.66e-01 | 1.00e+00 | 0.993 | 2 | 37 | 892 |
| GO:0000278 | mitotic cell cycle | 3.02e-01 | 1.00e+00 | 1.499 | 1 | 28 | 314 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 3.05e-01 | 1.00e+00 | 1.486 | 1 | 17 | 317 |
| GO:0005737 | cytoplasm | 3.42e-01 | 1.00e+00 | 0.431 | 4 | 127 | 2633 |
| GO:0005783 | endoplasmic reticulum | 3.55e-01 | 1.00e+00 | 1.220 | 1 | 12 | 381 |
| GO:0016020 | membrane | 4.02e-01 | 1.00e+00 | 0.557 | 2 | 56 | 1207 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 4.09e-01 | 1.00e+00 | 0.961 | 1 | 29 | 456 |
| GO:0008270 | zinc ion binding | 5.73e-01 | 1.00e+00 | 0.288 | 1 | 39 | 727 |
| GO:0070062 | extracellular vesicular exosome | 5.74e-01 | 1.00e+00 | 0.114 | 2 | 51 | 1641 |
| GO:0003677 | DNA binding | 6.75e-01 | 1.00e+00 | -0.093 | 1 | 52 | 947 |
| GO:0006351 | transcription, DNA-templated | 7.23e-01 | 1.00e+00 | -0.277 | 1 | 47 | 1076 |
| GO:0005634 | nucleus | 7.64e-01 | 1.00e+00 | -0.286 | 3 | 158 | 3246 |
| GO:0005730 | nucleolus | 7.69e-01 | 1.00e+00 | -0.455 | 1 | 74 | 1217 |