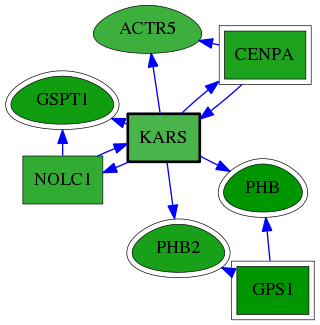
| GO:0000939 | condensed chromosome inner kinetochore | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0006430 | lysyl-tRNA aminoacylation | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0004824 | lysine-tRNA ligase activity | 8.19e-04 | 1.00e+00 | 10.253 | 1 | 1 | 1 |
| GO:0005515 | protein binding | 1.01e-03 | 1.00e+00 | 1.244 | 8 | 198 | 4124 |
| GO:0015966 | diadenosine tetraphosphate biosynthetic process | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 1 | 2 |
| GO:0003747 | translation release factor activity | 1.64e-03 | 1.00e+00 | 9.253 | 1 | 2 | 2 |
| GO:0000778 | condensed nuclear chromosome kinetochore | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 2 | 3 |
| GO:0071459 | protein localization to chromosome, centromeric region | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 3 |
| GO:0007000 | nucleolus organization | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 3.27e-03 | 1.00e+00 | 8.253 | 1 | 1 | 4 |
| GO:0007049 | cell cycle | 3.68e-03 | 1.00e+00 | 4.408 | 2 | 8 | 115 |
| GO:0060744 | mammary gland branching involved in thelarche | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 5 |
| GO:0050847 | progesterone receptor signaling pathway | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 2 | 6 |
| GO:0070914 | UV-damage excision repair | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 6 |
| GO:0000780 | condensed nuclear chromosome, centromeric region | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 1 | 7 |
| GO:0051382 | kinetochore assembly | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 2 | 7 |
| GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 5.72e-03 | 1.00e+00 | 7.446 | 1 | 2 | 7 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 1 | 8 |
| GO:0031011 | Ino80 complex | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 3 | 8 |
| GO:0010388 | cullin deneddylation | 6.54e-03 | 1.00e+00 | 7.253 | 1 | 4 | 8 |
| GO:0071354 | cellular response to interleukin-6 | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 9 |
| GO:0005095 | GTPase inhibitor activity | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 10 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 4 | 11 |
| GO:0006479 | protein methylation | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 2 | 11 |
| GO:0000132 | establishment of mitotic spindle orientation | 9.79e-03 | 1.00e+00 | 6.668 | 1 | 2 | 12 |
| GO:0005743 | mitochondrial inner membrane | 1.05e-02 | 1.00e+00 | 3.631 | 2 | 9 | 197 |
| GO:0005525 | GTP binding | 1.05e-02 | 1.00e+00 | 3.631 | 2 | 12 | 197 |
| GO:0060749 | mammary gland alveolus development | 1.06e-02 | 1.00e+00 | 6.553 | 1 | 1 | 13 |
| GO:0016597 | amino acid binding | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 1 | 14 |
| GO:0016575 | histone deacetylation | 1.14e-02 | 1.00e+00 | 6.446 | 1 | 2 | 14 |
| GO:0000188 | inactivation of MAPK activity | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 1 | 15 |
| GO:0005739 | mitochondrion | 1.33e-02 | 1.00e+00 | 2.474 | 3 | 32 | 659 |
| GO:0034080 | CENP-A containing nucleosome assembly | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 2 | 20 |
| GO:0071897 | DNA biosynthetic process | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 2 | 20 |
| GO:0030331 | estrogen receptor binding | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 2 | 22 |
| GO:0000049 | tRNA binding | 1.87e-02 | 1.00e+00 | 5.730 | 1 | 2 | 23 |
| GO:0005634 | nucleus | 1.93e-02 | 1.00e+00 | 1.174 | 6 | 158 | 3246 |
| GO:0008180 | COP9 signalosome | 1.95e-02 | 1.00e+00 | 5.668 | 1 | 6 | 24 |
| GO:0008033 | tRNA processing | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 2 | 25 |
| GO:0006418 | tRNA aminoacylation for protein translation | 2.35e-02 | 1.00e+00 | 5.395 | 1 | 4 | 29 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.58e-02 | 1.00e+00 | 2.945 | 2 | 17 | 317 |
| GO:0015030 | Cajal body | 2.59e-02 | 1.00e+00 | 5.253 | 1 | 3 | 32 |
| GO:0000786 | nucleosome | 2.75e-02 | 1.00e+00 | 5.166 | 1 | 2 | 34 |
| GO:0007254 | JNK cascade | 2.75e-02 | 1.00e+00 | 5.166 | 1 | 1 | 34 |
| GO:0000775 | chromosome, centromeric region | 3.31e-02 | 1.00e+00 | 4.896 | 1 | 6 | 41 |
| GO:0005737 | cytoplasm | 3.74e-02 | 1.00e+00 | 1.213 | 5 | 127 | 2633 |
| GO:0006310 | DNA recombination | 4.03e-02 | 1.00e+00 | 4.610 | 1 | 8 | 50 |
| GO:0006334 | nucleosome assembly | 4.10e-02 | 1.00e+00 | 4.581 | 1 | 4 | 51 |
| GO:0006302 | double-strand break repair | 4.18e-02 | 1.00e+00 | 4.553 | 1 | 5 | 52 |
| GO:0016032 | viral process | 4.50e-02 | 1.00e+00 | 2.512 | 2 | 30 | 428 |
| GO:0042826 | histone deacetylase binding | 4.66e-02 | 1.00e+00 | 4.395 | 1 | 5 | 58 |
| GO:0006415 | translational termination | 5.05e-02 | 1.00e+00 | 4.276 | 1 | 2 | 63 |
| GO:0006364 | rRNA processing | 5.36e-02 | 1.00e+00 | 4.187 | 1 | 2 | 67 |
| GO:0030308 | negative regulation of cell growth | 5.59e-02 | 1.00e+00 | 4.124 | 1 | 3 | 70 |
| GO:0016363 | nuclear matrix | 5.83e-02 | 1.00e+00 | 4.064 | 1 | 10 | 73 |
| GO:0001649 | osteoblast differentiation | 5.91e-02 | 1.00e+00 | 4.044 | 1 | 5 | 74 |
| GO:0015630 | microtubule cytoskeleton | 5.98e-02 | 1.00e+00 | 4.025 | 1 | 5 | 75 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6.68e-02 | 1.00e+00 | 3.861 | 1 | 5 | 84 |
| GO:0042981 | regulation of apoptotic process | 8.82e-02 | 1.00e+00 | 3.446 | 1 | 4 | 112 |
| GO:0003924 | GTPase activity | 9.35e-02 | 1.00e+00 | 3.359 | 1 | 7 | 119 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 9.80e-02 | 1.00e+00 | 3.288 | 1 | 4 | 125 |
| GO:0006184 | GTP catabolic process | 1.02e-01 | 1.00e+00 | 3.231 | 1 | 7 | 130 |
| GO:0005759 | mitochondrial matrix | 1.08e-01 | 1.00e+00 | 3.134 | 1 | 7 | 139 |
| GO:0044212 | transcription regulatory region DNA binding | 1.11e-01 | 1.00e+00 | 3.094 | 1 | 13 | 143 |
| GO:0006355 | regulation of transcription, DNA-templated | 1.12e-01 | 1.00e+00 | 1.772 | 2 | 31 | 715 |
| GO:0005622 | intracellular | 1.13e-01 | 1.00e+00 | 3.073 | 1 | 2 | 145 |
| GO:0007067 | mitotic nuclear division | 1.27e-01 | 1.00e+00 | 2.887 | 1 | 9 | 165 |
| GO:0044822 | poly(A) RNA binding | 1.35e-01 | 1.00e+00 | 1.611 | 2 | 45 | 799 |
| GO:0005654 | nucleoplasm | 1.57e-01 | 1.00e+00 | 1.479 | 2 | 68 | 876 |
| GO:0043234 | protein complex | 1.60e-01 | 1.00e+00 | 2.539 | 1 | 13 | 210 |
| GO:0019899 | enzyme binding | 1.60e-01 | 1.00e+00 | 2.539 | 1 | 7 | 210 |
| GO:0005524 | ATP binding | 1.61e-01 | 1.00e+00 | 1.453 | 2 | 37 | 892 |
| GO:0003682 | chromatin binding | 1.89e-01 | 1.00e+00 | 2.276 | 1 | 15 | 252 |
| GO:0046982 | protein heterodimerization activity | 1.98e-01 | 1.00e+00 | 2.204 | 1 | 11 | 265 |
| GO:0008285 | negative regulation of cell proliferation | 2.06e-01 | 1.00e+00 | 2.140 | 1 | 11 | 277 |
| GO:0006351 | transcription, DNA-templated | 2.18e-01 | 1.00e+00 | 1.182 | 2 | 47 | 1076 |
| GO:0000278 | mitotic cell cycle | 2.30e-01 | 1.00e+00 | 1.959 | 1 | 28 | 314 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 2.70e-01 | 1.00e+00 | 1.695 | 1 | 21 | 377 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.18e-01 | 1.00e+00 | 1.421 | 1 | 29 | 456 |
| GO:0010467 | gene expression | 3.63e-01 | 1.00e+00 | 1.190 | 1 | 49 | 535 |
| GO:0005887 | integral component of plasma membrane | 3.85e-01 | 1.00e+00 | 1.086 | 1 | 8 | 575 |
| GO:0070062 | extracellular vesicular exosome | 4.00e-01 | 1.00e+00 | 0.573 | 2 | 51 | 1641 |
| GO:0007165 | signal transduction | 4.07e-01 | 1.00e+00 | 0.982 | 1 | 24 | 618 |
| GO:0005576 | extracellular region | 4.30e-01 | 1.00e+00 | 0.881 | 1 | 15 | 663 |
| GO:0005829 | cytosol | 4.46e-01 | 1.00e+00 | 0.450 | 2 | 88 | 1787 |
| GO:0046872 | metal ion binding | 5.53e-01 | 1.00e+00 | 0.386 | 1 | 24 | 934 |
| GO:0003677 | DNA binding | 5.58e-01 | 1.00e+00 | 0.366 | 1 | 52 | 947 |
| GO:0016020 | membrane | 6.52e-01 | 1.00e+00 | 0.016 | 1 | 56 | 1207 |
| GO:0005730 | nucleolus | 6.55e-01 | 1.00e+00 | 0.004 | 1 | 74 | 1217 |
| GO:0005886 | plasma membrane | 8.01e-01 | 1.00e+00 | -0.547 | 1 | 46 | 1784 |