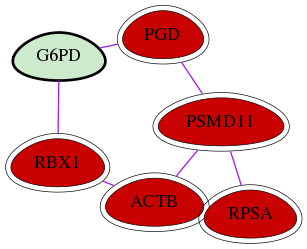
| GO:0019322 | pentose biosynthetic process | 1.44e-07 | 2.08e-03 | 11.231 | 2 | 2 | 2 |
| GO:0009051 | pentose-phosphate shunt, oxidative branch | 4.33e-07 | 6.24e-03 | 10.646 | 2 | 2 | 3 |
| GO:0006098 | pentose-phosphate shunt | 6.48e-06 | 9.35e-02 | 8.909 | 2 | 4 | 10 |
| GO:0005829 | cytosol | 2.67e-05 | 3.86e-01 | 2.531 | 6 | 132 | 2496 |
| GO:0050661 | NADP binding | 5.04e-05 | 7.27e-01 | 7.476 | 2 | 2 | 27 |
| GO:0021762 | substantia nigra development | 1.35e-04 | 1.00e+00 | 6.772 | 2 | 2 | 44 |
| GO:0046390 | ribose phosphate biosynthetic process | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0004345 | glucose-6-phosphate dehydrogenase activity | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0010734 | negative regulation of protein glutathionylation | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0019521 | D-gluconate metabolic process | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0070062 | extracellular vesicular exosome | 6.57e-04 | 1.00e+00 | 2.324 | 5 | 104 | 2400 |
| GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0005055 | laminin receptor activity | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0006740 | NADPH regeneration | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0006407 | rRNA export from nucleus | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0016032 | viral process | 9.28e-04 | 1.00e+00 | 3.755 | 3 | 55 | 534 |
| GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 4 |
| GO:0030686 | 90S preribosome | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 4 |
| GO:0031467 | Cul7-RING ubiquitin ligase complex | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 4 |
| GO:0019788 | NEDD8 ligase activity | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 4 |
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 3 | 5 |
| GO:0031461 | cullin-RING ubiquitin ligase complex | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 5 |
| GO:0043248 | proteasome assembly | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 5 |
| GO:0030891 | VCB complex | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 2 | 5 |
| GO:0016020 | membrane | 2.27e-03 | 1.00e+00 | 2.516 | 4 | 90 | 1681 |
| GO:0030957 | Tat protein binding | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 3 | 6 |
| GO:0006739 | NADP metabolic process | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 6 |
| GO:0031466 | Cul5-RING ubiquitin ligase complex | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 6 |
| GO:0051156 | glucose 6-phosphate metabolic process | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 7 |
| GO:0000028 | ribosomal small subunit assembly | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 7 |
| GO:0031462 | Cul2-RING ubiquitin ligase complex | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 2 | 7 |
| GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 2 | 8 |
| GO:0005536 | glucose binding | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 1 | 8 |
| GO:0070688 | MLL5-L complex | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 1 | 8 |
| GO:0045116 | protein neddylation | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 2 | 8 |
| GO:0016071 | mRNA metabolic process | 3.43e-03 | 1.00e+00 | 4.430 | 2 | 31 | 223 |
| GO:0016070 | RNA metabolic process | 4.19e-03 | 1.00e+00 | 4.283 | 2 | 32 | 247 |
| GO:0005975 | carbohydrate metabolic process | 4.39e-03 | 1.00e+00 | 4.248 | 2 | 9 | 253 |
| GO:0043249 | erythrocyte maturation | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 11 |
| GO:0005838 | proteasome regulatory particle | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 7 | 12 |
| GO:0035267 | NuA4 histone acetyltransferase complex | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 4 | 14 |
| GO:0032094 | response to food | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 15 |
| GO:0050998 | nitric-oxide synthase binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0022624 | proteasome accessory complex | 7.05e-03 | 1.00e+00 | 7.144 | 1 | 8 | 17 |
| GO:0005654 | nucleoplasm | 7.09e-03 | 1.00e+00 | 2.737 | 3 | 76 | 1082 |
| GO:0043523 | regulation of neuron apoptotic process | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 1 | 18 |
| GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 7.88e-03 | 1.00e+00 | 6.983 | 1 | 1 | 19 |
| GO:0048863 | stem cell differentiation | 7.88e-03 | 1.00e+00 | 6.983 | 1 | 1 | 19 |
| GO:0030863 | cortical cytoskeleton | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 1 | 22 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 4 | 22 |
| GO:0043044 | ATP-dependent chromatin remodeling | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 4 | 23 |
| GO:0031463 | Cul3-RING ubiquitin ligase complex | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 3 | 23 |
| GO:0043236 | laminin binding | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 1 | 23 |
| GO:0006513 | protein monoubiquitination | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 1 | 23 |
| GO:0044281 | small molecule metabolic process | 9.73e-03 | 1.00e+00 | 2.574 | 3 | 58 | 1211 |
| GO:0001816 | cytokine production | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 3 | 25 |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 3 | 26 |
| GO:0043022 | ribosome binding | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 3 | 27 |
| GO:0006749 | glutathione metabolic process | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 2 | 27 |
| GO:0031492 | nucleosomal DNA binding | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 4 | 27 |
| GO:0019894 | kinesin binding | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 1 | 28 |
| GO:0055114 | oxidation-reduction process | 1.17e-02 | 1.00e+00 | 3.517 | 2 | 12 | 420 |
| GO:0019005 | SCF ubiquitin ligase complex | 1.20e-02 | 1.00e+00 | 6.373 | 1 | 1 | 29 |
| GO:0034599 | cellular response to oxidative stress | 1.37e-02 | 1.00e+00 | 6.187 | 1 | 2 | 33 |
| GO:0001895 | retina homeostasis | 1.41e-02 | 1.00e+00 | 6.144 | 1 | 1 | 34 |
| GO:0006695 | cholesterol biosynthetic process | 1.41e-02 | 1.00e+00 | 6.144 | 1 | 4 | 34 |
| GO:0044267 | cellular protein metabolic process | 1.48e-02 | 1.00e+00 | 3.342 | 2 | 29 | 474 |
| GO:0034332 | adherens junction organization | 1.49e-02 | 1.00e+00 | 6.061 | 1 | 1 | 36 |
| GO:0051084 | 'de novo' posttranslational protein folding | 1.53e-02 | 1.00e+00 | 6.022 | 1 | 4 | 37 |
| GO:0070527 | platelet aggregation | 1.57e-02 | 1.00e+00 | 5.983 | 1 | 2 | 38 |
| GO:0022627 | cytosolic small ribosomal subunit | 1.61e-02 | 1.00e+00 | 5.946 | 1 | 4 | 39 |
| GO:0009898 | cytoplasmic side of plasma membrane | 1.69e-02 | 1.00e+00 | 5.874 | 1 | 1 | 41 |
| GO:0014070 | response to organic cyclic compound | 1.78e-02 | 1.00e+00 | 5.805 | 1 | 4 | 43 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 17 | 50 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 2.19e-02 | 1.00e+00 | 5.503 | 1 | 5 | 53 |
| GO:0045216 | cell-cell junction organization | 2.23e-02 | 1.00e+00 | 5.476 | 1 | 2 | 54 |
| GO:0000502 | proteasome complex | 2.39e-02 | 1.00e+00 | 5.373 | 1 | 17 | 58 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 19 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2.67e-02 | 1.00e+00 | 5.209 | 1 | 22 | 65 |
| GO:0034329 | cell junction assembly | 2.80e-02 | 1.00e+00 | 5.144 | 1 | 1 | 68 |
| GO:0010467 | gene expression | 2.85e-02 | 1.00e+00 | 2.845 | 2 | 59 | 669 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2.88e-02 | 1.00e+00 | 5.102 | 1 | 22 | 70 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2.96e-02 | 1.00e+00 | 5.061 | 1 | 20 | 72 |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 6 | 74 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 22 | 74 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 2 | 75 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 20 | 75 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 3.24e-02 | 1.00e+00 | 4.927 | 1 | 23 | 79 |
| GO:0019083 | viral transcription | 3.32e-02 | 1.00e+00 | 4.891 | 1 | 10 | 81 |
| GO:0045471 | response to ethanol | 3.53e-02 | 1.00e+00 | 4.805 | 1 | 3 | 86 |
| GO:0006415 | translational termination | 3.57e-02 | 1.00e+00 | 4.788 | 1 | 10 | 87 |
| GO:0006629 | lipid metabolic process | 3.69e-02 | 1.00e+00 | 4.739 | 1 | 5 | 90 |
| GO:0006928 | cellular component movement | 3.73e-02 | 1.00e+00 | 4.723 | 1 | 7 | 91 |
| GO:0005200 | structural constituent of cytoskeleton | 3.73e-02 | 1.00e+00 | 4.723 | 1 | 8 | 91 |
| GO:0071456 | cellular response to hypoxia | 3.77e-02 | 1.00e+00 | 4.708 | 1 | 6 | 92 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 3.77e-02 | 1.00e+00 | 4.708 | 1 | 21 | 92 |
| GO:0006414 | translational elongation | 3.81e-02 | 1.00e+00 | 4.692 | 1 | 13 | 93 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 4.25e-02 | 1.00e+00 | 4.531 | 1 | 10 | 104 |
| GO:0014069 | postsynaptic density | 4.29e-02 | 1.00e+00 | 4.517 | 1 | 3 | 105 |
| GO:0072562 | blood microparticle | 4.53e-02 | 1.00e+00 | 4.437 | 1 | 3 | 111 |
| GO:0030529 | ribonucleoprotein complex | 4.57e-02 | 1.00e+00 | 4.424 | 1 | 8 | 112 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 4.65e-02 | 1.00e+00 | 4.398 | 1 | 11 | 114 |
| GO:0019058 | viral life cycle | 4.69e-02 | 1.00e+00 | 4.386 | 1 | 13 | 115 |
| GO:0000209 | protein polyubiquitination | 4.73e-02 | 1.00e+00 | 4.373 | 1 | 20 | 116 |
| GO:0006325 | chromatin organization | 4.81e-02 | 1.00e+00 | 4.348 | 1 | 5 | 118 |
| GO:0007219 | Notch signaling pathway | 4.89e-02 | 1.00e+00 | 4.324 | 1 | 5 | 120 |
| GO:0005515 | protein binding | 4.97e-02 | 1.00e+00 | 0.997 | 5 | 184 | 6024 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 5.13e-02 | 1.00e+00 | 4.254 | 1 | 5 | 126 |
| GO:0000790 | nuclear chromatin | 5.25e-02 | 1.00e+00 | 4.220 | 1 | 9 | 129 |
| GO:0006413 | translational initiation | 5.33e-02 | 1.00e+00 | 4.198 | 1 | 17 | 131 |
| GO:0003735 | structural constituent of ribosome | 5.57e-02 | 1.00e+00 | 4.133 | 1 | 10 | 137 |
| GO:0006457 | protein folding | 5.80e-02 | 1.00e+00 | 4.071 | 1 | 7 | 143 |
| GO:0061024 | membrane organization | 5.88e-02 | 1.00e+00 | 4.051 | 1 | 7 | 145 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 6.04e-02 | 1.00e+00 | 4.012 | 1 | 32 | 149 |
| GO:0042981 | regulation of apoptotic process | 6.08e-02 | 1.00e+00 | 4.002 | 1 | 24 | 150 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 6.67e-02 | 1.00e+00 | 3.865 | 1 | 7 | 165 |
| GO:0030424 | axon | 6.71e-02 | 1.00e+00 | 3.856 | 1 | 4 | 166 |
| GO:0034641 | cellular nitrogen compound metabolic process | 6.91e-02 | 1.00e+00 | 3.813 | 1 | 20 | 171 |
| GO:0031625 | ubiquitin protein ligase binding | 7.18e-02 | 1.00e+00 | 3.755 | 1 | 14 | 178 |
| GO:0032403 | protein complex binding | 7.38e-02 | 1.00e+00 | 3.715 | 1 | 10 | 183 |
| GO:0005634 | nucleus | 8.39e-02 | 1.00e+00 | 1.077 | 4 | 136 | 4559 |
| GO:0006412 | translation | 9.20e-02 | 1.00e+00 | 3.386 | 1 | 20 | 230 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.02e-01 | 1.00e+00 | 3.237 | 1 | 6 | 255 |
| GO:0006281 | DNA repair | 1.04e-01 | 1.00e+00 | 3.203 | 1 | 18 | 261 |
| GO:0005856 | cytoskeleton | 1.15e-01 | 1.00e+00 | 3.046 | 1 | 12 | 291 |
| GO:0043234 | protein complex | 1.17e-01 | 1.00e+00 | 3.027 | 1 | 18 | 295 |
| GO:0016567 | protein ubiquitination | 1.17e-01 | 1.00e+00 | 3.027 | 1 | 5 | 295 |
| GO:0019901 | protein kinase binding | 1.25e-01 | 1.00e+00 | 2.923 | 1 | 21 | 317 |
| GO:0007411 | axon guidance | 1.26e-01 | 1.00e+00 | 2.914 | 1 | 13 | 319 |
| GO:0005925 | focal adhesion | 1.43e-01 | 1.00e+00 | 2.715 | 1 | 19 | 366 |
| GO:0007155 | cell adhesion | 1.44e-01 | 1.00e+00 | 2.704 | 1 | 6 | 369 |
| GO:0000278 | mitotic cell cycle | 1.52e-01 | 1.00e+00 | 2.620 | 1 | 48 | 391 |
| GO:0043066 | negative regulation of apoptotic process | 1.64e-01 | 1.00e+00 | 2.503 | 1 | 31 | 424 |
| GO:0007596 | blood coagulation | 1.75e-01 | 1.00e+00 | 2.401 | 1 | 18 | 455 |
| GO:0005737 | cytoplasm | 1.87e-01 | 1.00e+00 | 0.937 | 3 | 110 | 3767 |
| GO:0006915 | apoptotic process | 2.10e-01 | 1.00e+00 | 2.115 | 1 | 33 | 555 |
| GO:0042803 | protein homodimerization activity | 2.23e-01 | 1.00e+00 | 2.014 | 1 | 12 | 595 |
| GO:0045087 | innate immune response | 2.24e-01 | 1.00e+00 | 2.012 | 1 | 24 | 596 |
| GO:0005615 | extracellular space | 3.38e-01 | 1.00e+00 | 1.329 | 1 | 17 | 957 |
| GO:0008270 | zinc ion binding | 3.49e-01 | 1.00e+00 | 1.270 | 1 | 12 | 997 |
| GO:0044822 | poly(A) RNA binding | 3.66e-01 | 1.00e+00 | 1.187 | 1 | 49 | 1056 |
| GO:0005524 | ATP binding | 4.32e-01 | 1.00e+00 | 0.889 | 1 | 60 | 1298 |
| GO:0005886 | plasma membrane | 6.94e-01 | 1.00e+00 | -0.103 | 1 | 45 | 2582 |