int-snw-55775
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| chia-screen-data-Fav | 3.043 | 7.84e-18 | 3.17e-05 | 4.92e-04 |
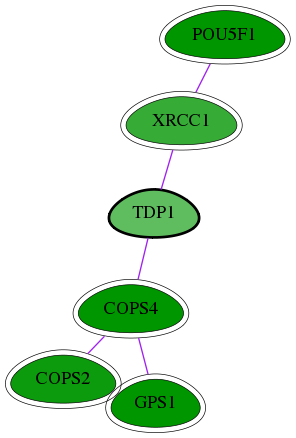
Genes (6)
| Gene Symbol | Entrez Gene ID | Frequency | chia-screen-data-Fav gene score | Best subnetwork score | Degree | Chia-Hits Primary | Chia-Hits Secondary |
|---|---|---|---|---|---|---|---|
| [ TDP1 ] | 55775 | 1 | -1.669 | 3.043 | 9 | - | - |
| POU5F1 | 5460 | 133 | -5.148 | 3.538 | 179 | Yes | Yes |
| COPS2 | 9318 | 2 | -2.531 | 3.043 | 46 | Yes | - |
| COPS4 | 51138 | 3 | -3.198 | 3.043 | 47 | Yes | Yes |
| GPS1 | 2873 | 3 | -3.621 | 3.043 | 26 | Yes | Yes |
| XRCC1 | 7515 | 3 | -2.092 | 3.043 | 34 | Yes | Yes |
Interactions (6)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| GPS1 | 2873 | COPS4 | 51138 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BIND, IntAct, MGI |
| COPS2 | 9318 | COPS4 | 51138 | pp | -- | int.I2D: BIND, HPRD, MGI; int.HPRD: in vitro, in vivo |
| COPS4 | 51138 | TDP1 | 55775 | pp | -- | int.I2D: BioGrid_Fly, BIND_Fly, FlyLow, MINT_Fly, IntAct_Fly |
| XRCC1 | 7515 | TDP1 | 55775 | pp | -- | int.I2D: MINT; int.Mint: MI:0915(physical association) |
| POU5F1 | 5460 | XRCC1 | 7515 | pp | -- | int.I2D: BioGrid_Mouse, IntAct_Mouse |
| GPS1 | 2873 | COPS2 | 9318 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct_Worm, MGI, MINT_Worm, BioGrid_Worm, BIND, BIND_Worm, CORE_1, HPRD, IntAct, INNATEDB; int.HPRD: in vitro, yeast 2-hybrid |
Related GO terms (66)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0010388 | cullin deneddylation | 3.36e-09 | 4.84e-05 | 9.646 | 3 | 5 | 9 |
| GO:0008180 | COP9 signalosome | 2.38e-07 | 3.44e-03 | 7.729 | 3 | 7 | 34 |
| GO:0000012 | single strand break repair | 3.03e-06 | 4.36e-02 | 9.424 | 2 | 3 | 7 |
| GO:0017005 | 3'-tyrosyl-DNA phosphodiesterase activity | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0060795 | cell fate commitment involved in formation of primary germ layer | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0090308 | regulation of methylation-dependent chromatin silencing | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0060965 | negative regulation of gene silencing by miRNA | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0009786 | regulation of asymmetric cell division | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0060913 | cardiac cell fate determination | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0003130 | BMP signaling pathway involved in heart induction | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0000338 | protein deneddylation | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0001714 | endodermal cell fate specification | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 5 |
| GO:0035413 | positive regulation of catenin import into nucleus | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 2 | 10 |
| GO:0060391 | positive regulation of SMAD protein import into nucleus | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 4 | 11 |
| GO:0006281 | DNA repair | 4.66e-03 | 1.00e+00 | 4.203 | 2 | 14 | 261 |
| GO:0035198 | miRNA binding | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 12 |
| GO:0005515 | protein binding | 5.30e-03 | 1.00e+00 | 1.260 | 6 | 198 | 6024 |
| GO:0004527 | exonuclease activity | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 1 | 13 |
| GO:0005095 | GTPase inhibitor activity | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 1 | 13 |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 2 | 13 |
| GO:0001824 | blastocyst development | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 2 | 15 |
| GO:0000188 | inactivation of MAPK activity | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 1 | 27 |
| GO:0010033 | response to organic substance | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 1 | 27 |
| GO:0006366 | transcription from RNA polymerase II promoter | 1.17e-02 | 1.00e+00 | 3.520 | 2 | 30 | 419 |
| GO:0005634 | nucleus | 1.39e-02 | 1.00e+00 | 1.399 | 5 | 159 | 4559 |
| GO:0006284 | base-excision repair | 1.53e-02 | 1.00e+00 | 6.022 | 1 | 3 | 37 |
| GO:0021766 | hippocampus development | 1.57e-02 | 1.00e+00 | 5.983 | 1 | 1 | 38 |
| GO:0035019 | somatic stem cell maintenance | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 6 | 40 |
| GO:0035914 | skeletal muscle cell differentiation | 1.69e-02 | 1.00e+00 | 5.874 | 1 | 2 | 41 |
| GO:0007254 | JNK cascade | 2.02e-02 | 1.00e+00 | 5.616 | 1 | 2 | 49 |
| GO:0003684 | damaged DNA binding | 2.02e-02 | 1.00e+00 | 5.616 | 1 | 2 | 49 |
| GO:0009611 | response to wounding | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 5 | 52 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 2.19e-02 | 1.00e+00 | 5.503 | 1 | 3 | 53 |
| GO:0006302 | double-strand break repair | 2.55e-02 | 1.00e+00 | 5.277 | 1 | 3 | 62 |
| GO:0010468 | regulation of gene expression | 2.80e-02 | 1.00e+00 | 5.144 | 1 | 2 | 68 |
| GO:0003697 | single-stranded DNA binding | 2.80e-02 | 1.00e+00 | 5.144 | 1 | 5 | 68 |
| GO:0008021 | synaptic vesicle | 3.00e-02 | 1.00e+00 | 5.041 | 1 | 2 | 73 |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 4 | 74 |
| GO:0030182 | neuron differentiation | 3.12e-02 | 1.00e+00 | 4.983 | 1 | 1 | 76 |
| GO:0009653 | anatomical structure morphogenesis | 3.40e-02 | 1.00e+00 | 4.856 | 1 | 2 | 83 |
| GO:0003690 | double-stranded DNA binding | 3.53e-02 | 1.00e+00 | 4.805 | 1 | 3 | 86 |
| GO:0005737 | cytoplasm | 4.38e-02 | 1.00e+00 | 1.352 | 4 | 124 | 3767 |
| GO:0001666 | response to hypoxia | 6.04e-02 | 1.00e+00 | 4.012 | 1 | 7 | 149 |
| GO:0044212 | transcription regulatory region DNA binding | 6.79e-02 | 1.00e+00 | 3.839 | 1 | 17 | 168 |
| GO:0005654 | nucleoplasm | 6.89e-02 | 1.00e+00 | 2.152 | 2 | 64 | 1082 |
| GO:0007049 | cell cycle | 7.02e-02 | 1.00e+00 | 3.788 | 1 | 6 | 174 |
| GO:0005667 | transcription factor complex | 7.06e-02 | 1.00e+00 | 3.780 | 1 | 17 | 175 |
| GO:0031625 | ubiquitin protein ligase binding | 7.18e-02 | 1.00e+00 | 3.755 | 1 | 5 | 178 |
| GO:0003714 | transcription corepressor activity | 7.18e-02 | 1.00e+00 | 3.755 | 1 | 11 | 178 |
| GO:0004871 | signal transducer activity | 8.35e-02 | 1.00e+00 | 3.531 | 1 | 4 | 208 |
| GO:0008134 | transcription factor binding | 9.66e-02 | 1.00e+00 | 3.312 | 1 | 18 | 242 |
| GO:0042493 | response to drug | 1.12e-01 | 1.00e+00 | 3.081 | 1 | 6 | 284 |
| GO:0008283 | cell proliferation | 1.29e-01 | 1.00e+00 | 2.874 | 1 | 14 | 328 |
| GO:0030054 | cell junction | 1.32e-01 | 1.00e+00 | 2.839 | 1 | 5 | 336 |
| GO:0043565 | sequence-specific DNA binding | 1.36e-01 | 1.00e+00 | 2.796 | 1 | 11 | 346 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.15e-01 | 1.00e+00 | 2.076 | 1 | 37 | 570 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 2.58e-01 | 1.00e+00 | 1.782 | 1 | 39 | 699 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 2.87e-01 | 1.00e+00 | 1.607 | 1 | 41 | 789 |
| GO:0007165 | signal transduction | 3.23e-01 | 1.00e+00 | 1.406 | 1 | 13 | 907 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.49e-01 | 1.00e+00 | 1.270 | 1 | 43 | 997 |
| GO:0044822 | poly(A) RNA binding | 3.66e-01 | 1.00e+00 | 1.187 | 1 | 50 | 1056 |
| GO:0003677 | DNA binding | 4.11e-01 | 1.00e+00 | 0.981 | 1 | 49 | 1218 |
| GO:0005730 | nucleolus | 5.16e-01 | 1.00e+00 | 0.551 | 1 | 66 | 1641 |
| GO:0070062 | extracellular vesicular exosome | 6.65e-01 | 1.00e+00 | 0.002 | 1 | 57 | 2400 |
| GO:0005829 | cytosol | 6.80e-01 | 1.00e+00 | -0.054 | 1 | 86 | 2496 |