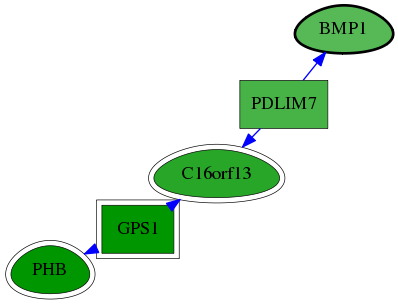
| GO:0001503 | ossification | 3.19e-04 | 1.00e+00 | 6.124 | 2 | 2 | 56 |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 5 |
| GO:0031988 | membrane-bounded vesicle | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 6 |
| GO:0050847 | progesterone receptor signaling pathway | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 2 | 6 |
| GO:0061036 | positive regulation of cartilage development | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 2 | 7 |
| GO:0010944 | negative regulation of transcription by competitive promoter binding | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 8 |
| GO:0010388 | cullin deneddylation | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 4 | 8 |
| GO:0030154 | cell differentiation | 4.17e-03 | 1.00e+00 | 4.259 | 2 | 5 | 204 |
| GO:0007275 | multicellular organismal development | 4.58e-03 | 1.00e+00 | 4.190 | 2 | 9 | 214 |
| GO:0071354 | cellular response to interleukin-6 | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 9 |
| GO:0005095 | GTPase inhibitor activity | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 10 |
| GO:0060766 | negative regulation of androgen receptor signaling pathway | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 4 | 11 |
| GO:0001502 | cartilage condensation | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 2 | 13 |
| GO:0016575 | histone deacetylation | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 2 | 14 |
| GO:0000188 | inactivation of MAPK activity | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 15 |
| GO:0071897 | DNA biosynthetic process | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 2 | 20 |
| GO:0008180 | COP9 signalosome | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 6 | 24 |
| GO:0008233 | peptidase activity | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 2 | 28 |
| GO:0042157 | lipoprotein metabolic process | 1.63e-02 | 1.00e+00 | 5.931 | 1 | 4 | 32 |
| GO:0007254 | JNK cascade | 1.73e-02 | 1.00e+00 | 5.844 | 1 | 1 | 34 |
| GO:0008237 | metallopeptidase activity | 1.93e-02 | 1.00e+00 | 5.684 | 1 | 3 | 38 |
| GO:0045669 | positive regulation of osteoblast differentiation | 2.08e-02 | 1.00e+00 | 5.574 | 1 | 2 | 41 |
| GO:0001725 | stress fiber | 2.33e-02 | 1.00e+00 | 5.408 | 1 | 6 | 46 |
| GO:0006898 | receptor-mediated endocytosis | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 1 | 52 |
| GO:0001726 | ruffle | 2.69e-02 | 1.00e+00 | 5.204 | 1 | 2 | 53 |
| GO:0042826 | histone deacetylase binding | 2.94e-02 | 1.00e+00 | 5.073 | 1 | 5 | 58 |
| GO:0004222 | metalloendopeptidase activity | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 1 | 64 |
| GO:0030308 | negative regulation of cell growth | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 3 | 70 |
| GO:0001649 | osteoblast differentiation | 3.73e-02 | 1.00e+00 | 4.722 | 1 | 5 | 74 |
| GO:0001501 | skeletal system development | 4.13e-02 | 1.00e+00 | 4.574 | 1 | 2 | 82 |
| GO:0022617 | extracellular matrix disassembly | 4.28e-02 | 1.00e+00 | 4.522 | 1 | 2 | 85 |
| GO:0008083 | growth factor activity | 4.38e-02 | 1.00e+00 | 4.489 | 1 | 1 | 87 |
| GO:0005125 | cytokine activity | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 1 | 89 |
| GO:0030036 | actin cytoskeleton organization | 4.53e-02 | 1.00e+00 | 4.440 | 1 | 4 | 90 |
| GO:0008270 | zinc ion binding | 4.76e-02 | 1.00e+00 | 2.426 | 2 | 39 | 727 |
| GO:0042981 | regulation of apoptotic process | 5.61e-02 | 1.00e+00 | 4.124 | 1 | 4 | 112 |
| GO:0005578 | proteinaceous extracellular matrix | 5.75e-02 | 1.00e+00 | 4.086 | 1 | 4 | 115 |
| GO:0007049 | cell cycle | 5.75e-02 | 1.00e+00 | 4.086 | 1 | 8 | 115 |
| GO:0015629 | actin cytoskeleton | 6.77e-02 | 1.00e+00 | 3.844 | 1 | 5 | 136 |
| GO:0044212 | transcription regulatory region DNA binding | 7.11e-02 | 1.00e+00 | 3.772 | 1 | 13 | 143 |
| GO:0005743 | mitochondrial inner membrane | 9.69e-02 | 1.00e+00 | 3.309 | 1 | 9 | 197 |
| GO:0019899 | enzyme binding | 1.03e-01 | 1.00e+00 | 3.217 | 1 | 7 | 210 |
| GO:0030198 | extracellular matrix organization | 1.08e-01 | 1.00e+00 | 3.144 | 1 | 7 | 221 |
| GO:0005737 | cytoplasm | 1.25e-01 | 1.00e+00 | 1.154 | 3 | 127 | 2633 |
| GO:0006508 | proteolysis | 1.28e-01 | 1.00e+00 | 2.887 | 1 | 11 | 264 |
| GO:0008285 | negative regulation of cell proliferation | 1.34e-01 | 1.00e+00 | 2.818 | 1 | 11 | 277 |
| GO:0005925 | focal adhesion | 1.36e-01 | 1.00e+00 | 2.792 | 1 | 14 | 282 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 1.52e-01 | 1.00e+00 | 2.623 | 1 | 17 | 317 |
| GO:0005509 | calcium ion binding | 1.67e-01 | 1.00e+00 | 2.476 | 1 | 9 | 351 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 1.79e-01 | 1.00e+00 | 2.373 | 1 | 21 | 377 |
| GO:0005794 | Golgi apparatus | 1.95e-01 | 1.00e+00 | 2.235 | 1 | 6 | 415 |
| GO:0005634 | nucleus | 2.08e-01 | 1.00e+00 | 0.852 | 3 | 158 | 3246 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.13e-01 | 1.00e+00 | 2.099 | 1 | 29 | 456 |
| GO:0005887 | integral component of plasma membrane | 2.62e-01 | 1.00e+00 | 1.764 | 1 | 8 | 575 |
| GO:0007165 | signal transduction | 2.79e-01 | 1.00e+00 | 1.660 | 1 | 24 | 618 |
| GO:0005615 | extracellular space | 2.92e-01 | 1.00e+00 | 1.585 | 1 | 15 | 651 |
| GO:0005739 | mitochondrion | 2.95e-01 | 1.00e+00 | 1.567 | 1 | 32 | 659 |
| GO:0005576 | extracellular region | 2.96e-01 | 1.00e+00 | 1.559 | 1 | 15 | 663 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.16e-01 | 1.00e+00 | 1.450 | 1 | 31 | 715 |
| GO:0005515 | protein binding | 3.57e-01 | 1.00e+00 | 0.507 | 3 | 198 | 4124 |
| GO:0044281 | small molecule metabolic process | 3.64e-01 | 1.00e+00 | 1.210 | 1 | 35 | 844 |
| GO:0005654 | nucleoplasm | 3.75e-01 | 1.00e+00 | 1.157 | 1 | 68 | 876 |
| GO:0016020 | membrane | 4.83e-01 | 1.00e+00 | 0.694 | 1 | 56 | 1207 |
| GO:0070062 | extracellular vesicular exosome | 6.02e-01 | 1.00e+00 | 0.251 | 1 | 51 | 1641 |