meta-reg-snw-8776
Interacting subnets
| Subnetwork | Dataset | Score | p-value 1 | p-value 2 | p-value 3 | Size | Highlighted genes |
|---|---|---|---|---|---|---|---|
| int-snw-6446 | wolf-screen-ratio-mammosphere-adherent | 0.930 | 2.55e-15 | 2.75e-03 | 4.33e-02 | 18 | 18 |
| reg-snw-8776 | wolf-screen-ratio-mammosphere-adherent | 0.800 | 2.67e-06 | 5.08e-03 | 9.11e-03 | 5 | 5 |
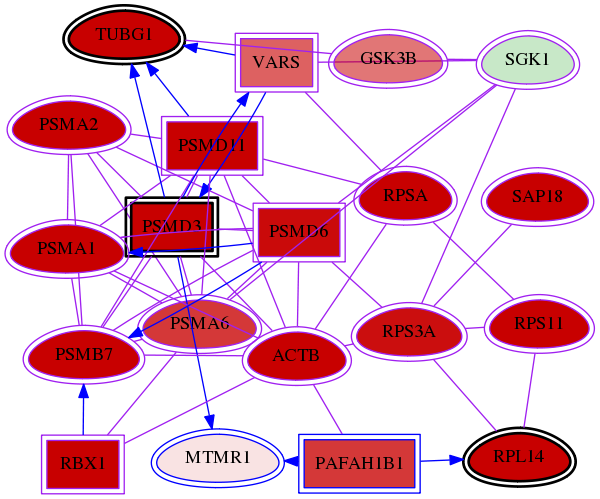
Genes (20)
| Gene Symbol | Entrez Gene ID | Frequency | wolf-screen-ratio-mammosphere-adherent gene score | Best subnetwork score | Degree | wolf adherent-list Hits GI | wolf mammosphere no adherent-list Hits GI |
|---|---|---|---|---|---|---|---|
| PSMA2 | 5683 | 112 | 1.093 | 1.106 | 108 | Yes | - |
| PAFAH1B1 | 5048 | 17 | 0.691 | 0.819 | 126 | Yes | - |
| RBX1 | 9978 | 115 | 1.185 | 0.934 | 148 | Yes | - |
| PSMB7 | 5695 | 118 | 0.982 | 0.934 | 90 | Yes | - |
| PSMA1 | 5682 | 100 | 0.996 | 0.878 | 152 | Yes | - |
| VARS | 7407 | 86 | 0.549 | 1.002 | 204 | Yes | - |
| RPSA | 3921 | 120 | 1.327 | 1.151 | 152 | Yes | - |
| RPS3A | 6189 | 40 | 0.835 | 1.069 | 166 | Yes | - |
| PSMA6 | 5687 | 19 | 0.691 | 0.956 | 137 | Yes | - |
| RPS11 | 6205 | 62 | 0.993 | 1.113 | 175 | Yes | - |
| PSMD3 | 5709 | 100 | 0.986 | 1.106 | 201 | Yes | - |
| RPL14 | 9045 | 49 | 1.250 | 1.113 | 166 | Yes | - |
| SGK1 | 6446 | 17 | -0.189 | 0.930 | 76 | - | Yes |
| GSK3B | 2932 | 22 | 0.475 | 0.934 | 319 | - | Yes |
| TUBG1 | 7283 | 98 | 0.974 | 0.973 | 91 | Yes | - |
| PSMD11 | 5717 | 124 | 1.095 | 1.106 | 218 | Yes | - |
| ACTB | 60 | 134 | 1.153 | 1.151 | 610 | Yes | - |
| SAP18 | 10284 | 20 | 1.115 | 1.069 | 57 | Yes | - |
| PSMD6 | 9861 | 79 | 0.848 | 0.878 | 143 | Yes | - |
| MTMR1 | 8776 | 2 | 0.097 | 0.800 | 49 | Yes | - |
Interactions (54)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| PSMD11 | 5717 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastHigh, BioGrid, INTEROLOG |
| SGK1 | 6446 | PSMD6 | 9861 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| PSMA6 | 5687 | RBX1 | 9978 | pp | -- | int.I2D: BioGrid |
| PSMB7 | 5695 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA2 | 5683 | PSMB7 | 5695 | pp | -- | int.Intact: MI:0914(association); int.I2D: BCI, BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct, Krogan_Core, MINT_Yeast, YeastLow, IntAct_Yeast |
| GSK3B | 2932 | SGK1 | 6446 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vitro, in vivo |
| PSMA2 | 5683 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| RPSA | 3921 | PSMD11 | 5717 | pp | -- | int.I2D: IntAct_Yeast |
| RPS3A | 6189 | PSMD6 | 9861 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| ACTB | 60 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA1 | 5682 | PSMD6 | 9861 | pd | < | reg.ITFP.txt: no annot |
| PSMA1 | 5682 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, YeastMedium |
| RPSA | 3921 | VARS | 7407 | pp | -- | int.I2D: IntAct_Yeast |
| PSMD3 | 5709 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastHigh, INTEROLOG |
| ACTB | 60 | RBX1 | 9978 | pp | -- | int.I2D: BioGrid_Yeast |
| GSK3B | 2932 | TUBG1 | 7283 | pp | -- | int.I2D: MINT |
| PSMA1 | 5682 | PSMA2 | 5683 | pp | -- | int.Intact: MI:0915(physical association), MI:0914(association); int.I2D: BioGrid_Worm, BioGrid_Yeast, BIND_Worm, CE_DATA, HPRD, IntAct, IntAct_Mouse, IntAct_Yeast, INTEROLOG, MINT_Worm, BioGrid, IntAct_Fly, IntAct_Worm, Krogan_Core, MINT_Yeast, YeastHigh; int.HPRD: yeast 2-hybrid |
| ACTB | 60 | RPSA | 3921 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct_Yeast, IntAct |
| TUBG1 | 7283 | VARS | 7407 | pd | < | reg.ITFP.txt: no annot |
| PAFAH1B1 | 5048 | RPL14 | 9045 | pd | > | reg.ITFP.txt: no annot |
| PSMA2 | 5683 | PSMA6 | 5687 | pp | -- | int.Intact: MI:0914(association), MI:0915(physical association); int.I2D: BCI, BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, HPRD, Krogan_Core, Tarassov_PCA, Yu_GoldStd; int.HPRD: yeast 2-hybrid |
| SGK1 | 6446 | VARS | 7407 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| RPS3A | 6189 | SAP18 | 10284 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, BioGrid, HPRD, MINT; int.Mint: MI:0915(physical association); int.HPRD: yeast 2-hybrid |
| PSMA1 | 5682 | PSMB7 | 5695 | pp | -- | int.Intact: MI:0914(association); int.I2D: BioGrid_Yeast, IntAct, IntAct_Mouse, IntAct_Yeast, YeastLow, Krogan_Core, MINT_Yeast |
| RPS3A | 6189 | RPL14 | 9045 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | PSMA1 | 5682 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMB7 | 5695 | RBX1 | 9978 | pd | < | reg.ITFP.txt: no annot |
| PSMD3 | 5709 | TUBG1 | 7283 | pd | > | reg.ITFP.txt: no annot |
| PSMB7 | 5695 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA6 | 5687 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| PSMD3 | 5709 | PSMD6 | 9861 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid_Worm, BioGrid_Yeast, BIND_Worm, CE_DATA, IntAct, IntAct_Yeast, MINT_Worm, MINT_Yeast, YeastHigh, BioGrid, BIND_Yeast, IntAct_Worm, INTEROLOG, Krogan_Core, MIPS |
| RPS3A | 6189 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | PAFAH1B1 | 5048 | pp | -- | int.I2D: BioGrid_Yeast |
| ACTB | 60 | PSMD6 | 9861 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS11 | 6205 | RPL14 | 9045 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMD3 | 5709 | VARS | 7407 | pd | <> | reg.ITFP.txt: no annot |
| ACTB | 60 | PSMA6 | 5687 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMA2 | 5683 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| PSMA1 | 5682 | PSMA6 | 5687 | pp | -- | int.Intact: MI:0914(association), MI:0915(physical association); int.I2D: BioGrid_Worm, BioGrid_Yeast, BIND_Worm, CE_DATA, IntAct, IntAct_Mouse, IntAct_Yeast, INTEROLOG, MINT_Worm, YeastHigh, IntAct_Worm, Krogan_Core, MINT_Yeast; int.Mint: MI:0914(association) |
| PSMD11 | 5717 | TUBG1 | 7283 | pd | > | reg.ITFP.txt: no annot |
| PSMA6 | 5687 | SGK1 | 6446 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| ACTB | 60 | PSMB7 | 5695 | pp | -- | int.I2D: BioGrid_Yeast |
| PSMB7 | 5695 | PSMD6 | 9861 | pd | < | reg.ITFP.txt: no annot |
| PSMB7 | 5695 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct |
| PAFAH1B1 | 5048 | MTMR1 | 8776 | pd | > | reg.ITFP.txt: no annot |
| ACTB | 60 | RPS3A | 6189 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| PSMA1 | 5682 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
| RPS3A | 6189 | SGK1 | 6446 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| PSMA6 | 5687 | PSMB7 | 5695 | pp | -- | int.I2D: BioGrid_Yeast, BIND_Yeast, Krogan_Core, MINT_Yeast, YeastLow |
| PSMA2 | 5683 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, YeastLow, BioGrid |
| PSMA6 | 5687 | PSMD6 | 9861 | pp | -- | int.Proteinpedia: Mass spectrometry; int.Intact: MI:0915(physical association); int.I2D: BioGrid_Yeast, IntAct, YeastLow |
| PSMD3 | 5709 | MTMR1 | 8776 | pd | > | reg.ITFP.txt: no annot |
| RPSA | 3921 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast |
| PSMA1 | 5682 | PSMD3 | 5709 | pp | -- | int.I2D: BioGrid_Yeast, YeastMedium |
| PSMA6 | 5687 | PSMD11 | 5717 | pp | -- | int.I2D: BioGrid_Yeast, YeastLow |
Related GO terms (331)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0016071 | mRNA metabolic process | 3.65e-16 | 5.95e-12 | 5.331 | 11 | 34 | 223 |
| GO:0016070 | RNA metabolic process | 1.14e-15 | 1.86e-11 | 5.184 | 11 | 34 | 247 |
| GO:0005829 | cytosol | 8.47e-15 | 1.38e-10 | 2.597 | 19 | 125 | 2562 |
| GO:0006521 | regulation of cellular amino acid metabolic process | 1.23e-13 | 2.01e-09 | 6.836 | 7 | 21 | 50 |
| GO:0016032 | viral process | 1.51e-13 | 2.46e-09 | 4.181 | 12 | 55 | 540 |
| GO:0000502 | proteasome complex | 3.68e-13 | 6.01e-09 | 6.622 | 7 | 22 | 58 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 8.48e-13 | 1.38e-08 | 6.458 | 7 | 24 | 65 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 8.48e-13 | 1.38e-08 | 6.458 | 7 | 22 | 65 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1.45e-12 | 2.37e-08 | 6.351 | 7 | 24 | 70 |
| GO:0010467 | gene expression | 1.90e-12 | 3.11e-08 | 3.872 | 12 | 58 | 669 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 1.97e-12 | 3.22e-08 | 6.290 | 7 | 23 | 73 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2.18e-12 | 3.55e-08 | 6.270 | 7 | 24 | 74 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 2.90e-12 | 4.74e-08 | 6.213 | 7 | 23 | 77 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 3.50e-12 | 5.71e-08 | 6.176 | 7 | 25 | 79 |
| GO:0042981 | regulation of apoptotic process | 5.11e-12 | 8.34e-08 | 5.434 | 8 | 26 | 151 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 1.22e-11 | 1.99e-07 | 5.925 | 7 | 23 | 94 |
| GO:0000209 | protein polyubiquitination | 5.48e-11 | 8.94e-07 | 5.622 | 7 | 21 | 116 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 3.37e-10 | 5.51e-06 | 5.251 | 7 | 33 | 150 |
| GO:0000278 | mitotic cell cycle | 3.69e-10 | 6.03e-06 | 4.206 | 9 | 52 | 398 |
| GO:0043066 | negative regulation of apoptotic process | 7.77e-10 | 1.27e-05 | 4.084 | 9 | 30 | 433 |
| GO:0034641 | cellular nitrogen compound metabolic process | 1.08e-09 | 1.76e-05 | 5.012 | 7 | 25 | 177 |
| GO:0005839 | proteasome core complex | 4.96e-09 | 8.10e-05 | 7.503 | 4 | 11 | 18 |
| GO:0004298 | threonine-type endopeptidase activity | 7.84e-09 | 1.28e-04 | 7.351 | 4 | 11 | 20 |
| GO:0019773 | proteasome core complex, alpha-subunit complex | 8.78e-08 | 1.43e-03 | 8.257 | 3 | 5 | 8 |
| GO:0006915 | apoptotic process | 1.86e-07 | 3.03e-03 | 3.515 | 8 | 34 | 571 |
| GO:0070062 | extracellular vesicular exosome | 7.17e-07 | 1.17e-02 | 2.076 | 13 | 98 | 2516 |
| GO:0022624 | proteasome accessory complex | 1.06e-06 | 1.73e-02 | 7.170 | 3 | 9 | 17 |
| GO:0005654 | nucleoplasm | 2.28e-06 | 3.72e-02 | 2.746 | 9 | 83 | 1095 |
| GO:0019083 | viral transcription | 2.57e-06 | 4.19e-02 | 5.333 | 4 | 8 | 81 |
| GO:0006415 | translational termination | 3.42e-06 | 5.58e-02 | 5.230 | 4 | 8 | 87 |
| GO:0006414 | translational elongation | 4.46e-06 | 7.28e-02 | 5.133 | 4 | 11 | 93 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 6.97e-06 | 1.14e-01 | 4.972 | 4 | 8 | 104 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1.00e-05 | 1.64e-01 | 4.840 | 4 | 10 | 114 |
| GO:0019058 | viral life cycle | 1.04e-05 | 1.70e-01 | 4.827 | 4 | 10 | 115 |
| GO:0022627 | cytosolic small ribosomal subunit | 1.40e-05 | 2.28e-01 | 5.972 | 3 | 3 | 39 |
| GO:0006413 | translational initiation | 1.74e-05 | 2.84e-01 | 4.639 | 4 | 12 | 131 |
| GO:0003735 | structural constituent of ribosome | 2.32e-05 | 3.79e-01 | 4.533 | 4 | 8 | 141 |
| GO:0044281 | small molecule metabolic process | 8.14e-05 | 1.00e+00 | 2.334 | 8 | 57 | 1295 |
| GO:0005838 | proteasome regulatory particle | 9.35e-05 | 1.00e+00 | 7.088 | 2 | 7 | 12 |
| GO:0006412 | translation | 1.69e-04 | 1.00e+00 | 3.796 | 4 | 15 | 235 |
| GO:0044267 | cellular protein metabolic process | 2.67e-04 | 1.00e+00 | 3.043 | 5 | 24 | 495 |
| GO:0030529 | ribonucleoprotein complex | 3.47e-04 | 1.00e+00 | 4.425 | 3 | 8 | 114 |
| GO:0005844 | polysome | 4.21e-04 | 1.00e+00 | 6.029 | 2 | 4 | 25 |
| GO:0051059 | NF-kappaB binding | 4.21e-04 | 1.00e+00 | 6.029 | 2 | 3 | 25 |
| GO:0005813 | centrosome | 6.81e-04 | 1.00e+00 | 3.267 | 4 | 12 | 339 |
| GO:0005515 | protein binding | 7.38e-04 | 1.00e+00 | 0.998 | 15 | 172 | 6127 |
| GO:0003723 | RNA binding | 8.09e-04 | 1.00e+00 | 3.201 | 4 | 19 | 355 |
| GO:0021766 | hippocampus development | 1.03e-03 | 1.00e+00 | 5.387 | 2 | 4 | 39 |
| GO:0000235 | astral microtubule | 1.23e-03 | 1.00e+00 | 9.673 | 1 | 1 | 1 |
| GO:0051660 | establishment of centrosome localization | 1.23e-03 | 1.00e+00 | 9.673 | 1 | 1 | 1 |
| GO:0035639 | purine ribonucleoside triphosphate binding | 1.23e-03 | 1.00e+00 | 9.673 | 1 | 1 | 1 |
| GO:0060453 | regulation of gastric acid secretion | 1.23e-03 | 1.00e+00 | 9.673 | 1 | 1 | 1 |
| GO:0046469 | platelet activating factor metabolic process | 1.23e-03 | 1.00e+00 | 9.673 | 1 | 1 | 1 |
| GO:0000226 | microtubule cytoskeleton organization | 2.04e-03 | 1.00e+00 | 4.891 | 2 | 3 | 55 |
| GO:0000932 | cytoplasmic mRNA processing body | 2.11e-03 | 1.00e+00 | 4.865 | 2 | 3 | 56 |
| GO:0004832 | valine-tRNA ligase activity | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0051081 | nuclear envelope disassembly | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 2 | 2 |
| GO:0006407 | rRNA export from nucleus | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0006438 | valyl-tRNA aminoacylation | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0036035 | osteoclast development | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 2 | 2 |
| GO:0071109 | superior temporal gyrus development | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:2000077 | negative regulation of type B pancreatic cell development | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0005055 | laminin receptor activity | 2.45e-03 | 1.00e+00 | 8.673 | 1 | 1 | 2 |
| GO:0005737 | cytoplasm | 3.15e-03 | 1.00e+00 | 1.175 | 11 | 98 | 3976 |
| GO:0061574 | ASAP complex | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:0014043 | negative regulation of neuron maturation | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:0000320 | re-entry into mitotic cell cycle | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:2000466 | negative regulation of glycogen (starch) synthase activity | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:0044027 | hypermethylation of CpG island | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:0021540 | corpus callosum morphogenesis | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:0045505 | dynein intermediate chain binding | 3.67e-03 | 1.00e+00 | 8.088 | 1 | 1 | 3 |
| GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0000212 | meiotic spindle organization | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0030686 | 90S preribosome | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0070294 | renal sodium ion absorption | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0019788 | NEDD8 ligase activity | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0031467 | Cul7-RING ubiquitin ligase complex | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0051534 | negative regulation of NFAT protein import into nucleus | 4.89e-03 | 1.00e+00 | 7.673 | 1 | 1 | 4 |
| GO:0005200 | structural constituent of cytoskeleton | 5.71e-03 | 1.00e+00 | 4.133 | 2 | 7 | 93 |
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 2 | 5 |
| GO:0031023 | microtubule organizing center organization | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0031461 | cullin-RING ubiquitin ligase complex | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0043248 | proteasome assembly | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0036016 | cellular response to interleukin-3 | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:2000738 | positive regulation of stem cell differentiation | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0030891 | VCB complex | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 2 | 5 |
| GO:0017081 | chloride channel regulator activity | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0005827 | polar microtubule | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0032411 | positive regulation of transporter activity | 6.11e-03 | 1.00e+00 | 7.351 | 1 | 1 | 5 |
| GO:0030426 | growth cone | 6.20e-03 | 1.00e+00 | 4.073 | 2 | 3 | 97 |
| GO:0002181 | cytoplasmic translation | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0001667 | ameboidal-type cell migration | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0031466 | Cul5-RING ubiquitin ligase complex | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0008090 | retrograde axon cargo transport | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0050774 | negative regulation of dendrite morphogenesis | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0030957 | Tat protein binding | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 4 | 6 |
| GO:0034452 | dynactin binding | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0010614 | negative regulation of cardiac muscle hypertrophy | 7.33e-03 | 1.00e+00 | 7.088 | 1 | 1 | 6 |
| GO:0044822 | poly(A) RNA binding | 8.38e-03 | 1.00e+00 | 1.920 | 5 | 50 | 1078 |
| GO:0007097 | nuclear migration | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 1 | 7 |
| GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 1 | 7 |
| GO:0000028 | ribosomal small subunit assembly | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 1 | 7 |
| GO:0002161 | aminoacyl-tRNA editing activity | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 2 | 7 |
| GO:0031462 | Cul2-RING ubiquitin ligase complex | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 2 | 7 |
| GO:0017145 | stem cell division | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 1 | 7 |
| GO:0000930 | gamma-tubulin complex | 8.55e-03 | 1.00e+00 | 6.865 | 1 | 1 | 7 |
| GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 2 | 8 |
| GO:0004438 | phosphatidylinositol-3-phosphatase activity | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 1 | 8 |
| GO:0070688 | MLL5-L complex | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 1 | 8 |
| GO:0045116 | protein neddylation | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 2 | 8 |
| GO:0031512 | motile primary cilium | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 1 | 8 |
| GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 1 | 8 |
| GO:0045719 | negative regulation of glycogen biosynthetic process | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 1 | 8 |
| GO:0035372 | protein localization to microtubule | 9.76e-03 | 1.00e+00 | 6.673 | 1 | 1 | 8 |
| GO:0005925 | focal adhesion | 9.89e-03 | 1.00e+00 | 2.726 | 3 | 18 | 370 |
| GO:0048156 | tau protein binding | 1.10e-02 | 1.00e+00 | 6.503 | 1 | 1 | 9 |
| GO:0006983 | ER overload response | 1.10e-02 | 1.00e+00 | 6.503 | 1 | 2 | 9 |
| GO:0032319 | regulation of Rho GTPase activity | 1.10e-02 | 1.00e+00 | 6.503 | 1 | 2 | 9 |
| GO:0021895 | cerebral cortex neuron differentiation | 1.10e-02 | 1.00e+00 | 6.503 | 1 | 1 | 9 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 1.20e-02 | 1.00e+00 | 3.574 | 2 | 7 | 137 |
| GO:0006450 | regulation of translational fidelity | 1.22e-02 | 1.00e+00 | 6.351 | 1 | 2 | 10 |
| GO:0010226 | response to lithium ion | 1.22e-02 | 1.00e+00 | 6.351 | 1 | 2 | 10 |
| GO:0001675 | acrosome assembly | 1.22e-02 | 1.00e+00 | 6.351 | 1 | 1 | 10 |
| GO:0030877 | beta-catenin destruction complex | 1.22e-02 | 1.00e+00 | 6.351 | 1 | 2 | 10 |
| GO:0005730 | nucleolus | 1.30e-02 | 1.00e+00 | 1.540 | 6 | 70 | 1684 |
| GO:0045502 | dynein binding | 1.34e-02 | 1.00e+00 | 6.213 | 1 | 2 | 11 |
| GO:0021819 | layer formation in cerebral cortex | 1.34e-02 | 1.00e+00 | 6.213 | 1 | 1 | 11 |
| GO:0032886 | regulation of microtubule-based process | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 4 | 12 |
| GO:0034236 | protein kinase A catalytic subunit binding | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 12 |
| GO:0047496 | vesicle transport along microtubule | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 12 |
| GO:0050321 | tau-protein kinase activity | 1.46e-02 | 1.00e+00 | 6.088 | 1 | 1 | 12 |
| GO:0005634 | nucleus | 1.53e-02 | 1.00e+00 | 0.895 | 11 | 131 | 4828 |
| GO:0016020 | membrane | 1.54e-02 | 1.00e+00 | 1.488 | 6 | 80 | 1746 |
| GO:0042273 | ribosomal large subunit biogenesis | 1.58e-02 | 1.00e+00 | 5.972 | 1 | 4 | 13 |
| GO:0030234 | enzyme regulator activity | 1.58e-02 | 1.00e+00 | 5.972 | 1 | 3 | 13 |
| GO:0046827 | positive regulation of protein export from nucleus | 1.58e-02 | 1.00e+00 | 5.972 | 1 | 2 | 13 |
| GO:0035267 | NuA4 histone acetyltransferase complex | 1.70e-02 | 1.00e+00 | 5.865 | 1 | 4 | 14 |
| GO:0031333 | negative regulation of protein complex assembly | 1.70e-02 | 1.00e+00 | 5.865 | 1 | 1 | 14 |
| GO:0048168 | regulation of neuronal synaptic plasticity | 1.70e-02 | 1.00e+00 | 5.865 | 1 | 1 | 14 |
| GO:0007020 | microtubule nucleation | 1.70e-02 | 1.00e+00 | 5.865 | 1 | 1 | 14 |
| GO:0031334 | positive regulation of protein complex assembly | 1.70e-02 | 1.00e+00 | 5.865 | 1 | 2 | 14 |
| GO:0006349 | regulation of gene expression by genetic imprinting | 1.82e-02 | 1.00e+00 | 5.766 | 1 | 1 | 15 |
| GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 1.82e-02 | 1.00e+00 | 5.766 | 1 | 1 | 15 |
| GO:0048854 | brain morphogenesis | 1.82e-02 | 1.00e+00 | 5.766 | 1 | 1 | 15 |
| GO:0030424 | axon | 1.85e-02 | 1.00e+00 | 3.246 | 2 | 3 | 172 |
| GO:0007405 | neuroblast proliferation | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 1 | 16 |
| GO:0042176 | regulation of protein catabolic process | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 3 | 16 |
| GO:0007520 | myoblast fusion | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 1 | 16 |
| GO:0046856 | phosphatidylinositol dephosphorylation | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 1 | 16 |
| GO:0051603 | proteolysis involved in cellular protein catabolic process | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 2 | 16 |
| GO:0050998 | nitric-oxide synthase binding | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 1 | 16 |
| GO:0019226 | transmission of nerve impulse | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 1 | 16 |
| GO:0000132 | establishment of mitotic spindle orientation | 1.94e-02 | 1.00e+00 | 5.673 | 1 | 1 | 16 |
| GO:0031625 | ubiquitin protein ligase binding | 2.02e-02 | 1.00e+00 | 3.181 | 2 | 13 | 180 |
| GO:0035255 | ionotropic glutamate receptor binding | 2.06e-02 | 1.00e+00 | 5.585 | 1 | 1 | 17 |
| GO:0043274 | phospholipase binding | 2.06e-02 | 1.00e+00 | 5.585 | 1 | 1 | 17 |
| GO:0031122 | cytoplasmic microtubule organization | 2.18e-02 | 1.00e+00 | 5.503 | 1 | 2 | 18 |
| GO:0005246 | calcium channel regulator activity | 2.18e-02 | 1.00e+00 | 5.503 | 1 | 1 | 18 |
| GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 2.18e-02 | 1.00e+00 | 5.503 | 1 | 1 | 18 |
| GO:0045773 | positive regulation of axon extension | 2.18e-02 | 1.00e+00 | 5.503 | 1 | 2 | 18 |
| GO:0035145 | exon-exon junction complex | 2.30e-02 | 1.00e+00 | 5.425 | 1 | 2 | 19 |
| GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 2.30e-02 | 1.00e+00 | 5.425 | 1 | 1 | 19 |
| GO:0048863 | stem cell differentiation | 2.30e-02 | 1.00e+00 | 5.425 | 1 | 1 | 19 |
| GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity | 2.42e-02 | 1.00e+00 | 5.351 | 1 | 1 | 20 |
| GO:0009306 | protein secretion | 2.54e-02 | 1.00e+00 | 5.280 | 1 | 1 | 21 |
| GO:0001954 | positive regulation of cell-matrix adhesion | 2.54e-02 | 1.00e+00 | 5.280 | 1 | 1 | 21 |
| GO:0030010 | establishment of cell polarity | 2.54e-02 | 1.00e+00 | 5.280 | 1 | 2 | 21 |
| GO:0017080 | sodium channel regulator activity | 2.66e-02 | 1.00e+00 | 5.213 | 1 | 1 | 22 |
| GO:0030863 | cortical cytoskeleton | 2.66e-02 | 1.00e+00 | 5.213 | 1 | 1 | 22 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 2.66e-02 | 1.00e+00 | 5.213 | 1 | 4 | 22 |
| GO:0031463 | Cul3-RING ubiquitin ligase complex | 2.78e-02 | 1.00e+00 | 5.149 | 1 | 2 | 23 |
| GO:0043236 | laminin binding | 2.78e-02 | 1.00e+00 | 5.149 | 1 | 1 | 23 |
| GO:0006513 | protein monoubiquitination | 2.78e-02 | 1.00e+00 | 5.149 | 1 | 1 | 23 |
| GO:0043044 | ATP-dependent chromatin remodeling | 2.78e-02 | 1.00e+00 | 5.149 | 1 | 4 | 23 |
| GO:0000794 | condensed nuclear chromosome | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 2 | 24 |
| GO:0046329 | negative regulation of JNK cascade | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 24 |
| GO:0005977 | glycogen metabolic process | 2.90e-02 | 1.00e+00 | 5.088 | 1 | 1 | 24 |
| GO:0006611 | protein export from nucleus | 3.02e-02 | 1.00e+00 | 5.029 | 1 | 4 | 25 |
| GO:0030016 | myofibril | 3.02e-02 | 1.00e+00 | 5.029 | 1 | 1 | 25 |
| GO:0045931 | positive regulation of mitotic cell cycle | 3.14e-02 | 1.00e+00 | 4.972 | 1 | 1 | 26 |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 3.14e-02 | 1.00e+00 | 4.972 | 1 | 2 | 26 |
| GO:0015459 | potassium channel regulator activity | 3.14e-02 | 1.00e+00 | 4.972 | 1 | 1 | 26 |
| GO:0007616 | long-term memory | 3.26e-02 | 1.00e+00 | 4.918 | 1 | 1 | 27 |
| GO:0019843 | rRNA binding | 3.26e-02 | 1.00e+00 | 4.918 | 1 | 3 | 27 |
| GO:0005875 | microtubule associated complex | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 2 | 28 |
| GO:0007017 | microtubule-based process | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 3 | 28 |
| GO:0043022 | ribosome binding | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 3 | 28 |
| GO:0031492 | nucleosomal DNA binding | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 4 | 28 |
| GO:0000118 | histone deacetylase complex | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 1 | 28 |
| GO:0019894 | kinesin binding | 3.38e-02 | 1.00e+00 | 4.865 | 1 | 1 | 28 |
| GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 3.50e-02 | 1.00e+00 | 4.815 | 1 | 2 | 29 |
| GO:0031252 | cell leading edge | 3.50e-02 | 1.00e+00 | 4.815 | 1 | 3 | 29 |
| GO:0019005 | SCF ubiquitin ligase complex | 3.50e-02 | 1.00e+00 | 4.815 | 1 | 1 | 29 |
| GO:0004712 | protein serine/threonine/tyrosine kinase activity | 3.50e-02 | 1.00e+00 | 4.815 | 1 | 1 | 29 |
| GO:0043198 | dendritic shaft | 3.50e-02 | 1.00e+00 | 4.815 | 1 | 1 | 29 |
| GO:0010977 | negative regulation of neuron projection development | 3.61e-02 | 1.00e+00 | 4.766 | 1 | 2 | 30 |
| GO:0043025 | neuronal cell body | 3.81e-02 | 1.00e+00 | 2.684 | 2 | 4 | 254 |
| GO:0051219 | phosphoprotein binding | 3.85e-02 | 1.00e+00 | 4.673 | 1 | 3 | 32 |
| GO:1903507 | negative regulation of nucleic acid-templated transcription | 3.85e-02 | 1.00e+00 | 4.673 | 1 | 2 | 32 |
| GO:0032091 | negative regulation of protein binding | 3.97e-02 | 1.00e+00 | 4.628 | 1 | 1 | 33 |
| GO:0001837 | epithelial to mesenchymal transition | 3.97e-02 | 1.00e+00 | 4.628 | 1 | 1 | 33 |
| GO:0043407 | negative regulation of MAP kinase activity | 4.09e-02 | 1.00e+00 | 4.585 | 1 | 1 | 34 |
| GO:0007611 | learning or memory | 4.09e-02 | 1.00e+00 | 4.585 | 1 | 2 | 34 |
| GO:0030017 | sarcomere | 4.09e-02 | 1.00e+00 | 4.585 | 1 | 1 | 34 |
| GO:0001085 | RNA polymerase II transcription factor binding | 4.09e-02 | 1.00e+00 | 4.585 | 1 | 2 | 34 |
| GO:0004175 | endopeptidase activity | 4.09e-02 | 1.00e+00 | 4.585 | 1 | 2 | 34 |
| GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 4.21e-02 | 1.00e+00 | 4.543 | 1 | 2 | 35 |
| GO:0034332 | adherens junction organization | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 36 |
| GO:0032855 | positive regulation of Rac GTPase activity | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 36 |
| GO:0001895 | retina homeostasis | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 1 | 36 |
| GO:0016301 | kinase activity | 4.44e-02 | 1.00e+00 | 4.463 | 1 | 3 | 37 |
| GO:0051084 | 'de novo' posttranslational protein folding | 4.44e-02 | 1.00e+00 | 4.463 | 1 | 4 | 37 |
| GO:0070527 | platelet aggregation | 4.56e-02 | 1.00e+00 | 4.425 | 1 | 2 | 38 |
| GO:0032092 | positive regulation of protein binding | 4.67e-02 | 1.00e+00 | 4.387 | 1 | 3 | 39 |
| GO:0050885 | neuromuscular process controlling balance | 4.91e-02 | 1.00e+00 | 4.315 | 1 | 1 | 41 |
| GO:0006418 | tRNA aminoacylation for protein translation | 5.03e-02 | 1.00e+00 | 4.280 | 1 | 5 | 42 |
| GO:0021987 | cerebral cortex development | 5.03e-02 | 1.00e+00 | 4.280 | 1 | 3 | 42 |
| GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 5.26e-02 | 1.00e+00 | 4.213 | 1 | 3 | 44 |
| GO:0005871 | kinesin complex | 5.26e-02 | 1.00e+00 | 4.213 | 1 | 2 | 44 |
| GO:0021762 | substantia nigra development | 5.49e-02 | 1.00e+00 | 4.149 | 1 | 1 | 46 |
| GO:0004674 | protein serine/threonine kinase activity | 5.52e-02 | 1.00e+00 | 2.387 | 2 | 6 | 312 |
| GO:0050727 | regulation of inflammatory response | 5.61e-02 | 1.00e+00 | 4.118 | 1 | 3 | 47 |
| GO:0008344 | adult locomotory behavior | 5.61e-02 | 1.00e+00 | 4.118 | 1 | 1 | 47 |
| GO:0006950 | response to stress | 5.72e-02 | 1.00e+00 | 4.088 | 1 | 3 | 48 |
| GO:0019901 | protein kinase binding | 5.77e-02 | 1.00e+00 | 2.351 | 2 | 21 | 320 |
| GO:0022625 | cytosolic large ribosomal subunit | 5.84e-02 | 1.00e+00 | 4.058 | 1 | 5 | 49 |
| GO:0007411 | axon guidance | 6.00e-02 | 1.00e+00 | 2.319 | 2 | 9 | 327 |
| GO:0045444 | fat cell differentiation | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 1 | 51 |
| GO:0045732 | positive regulation of protein catabolic process | 6.07e-02 | 1.00e+00 | 4.000 | 1 | 4 | 51 |
| GO:0016042 | lipid catabolic process | 6.19e-02 | 1.00e+00 | 3.972 | 1 | 1 | 52 |
| GO:0030334 | regulation of cell migration | 6.19e-02 | 1.00e+00 | 3.972 | 1 | 2 | 52 |
| GO:0007623 | circadian rhythm | 6.42e-02 | 1.00e+00 | 3.918 | 1 | 1 | 54 |
| GO:0006661 | phosphatidylinositol biosynthetic process | 6.42e-02 | 1.00e+00 | 3.918 | 1 | 1 | 54 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 6.42e-02 | 1.00e+00 | 3.918 | 1 | 5 | 54 |
| GO:0002039 | p53 binding | 6.53e-02 | 1.00e+00 | 3.891 | 1 | 7 | 55 |
| GO:0006814 | sodium ion transport | 6.53e-02 | 1.00e+00 | 3.891 | 1 | 1 | 55 |
| GO:0008217 | regulation of blood pressure | 6.88e-02 | 1.00e+00 | 3.815 | 1 | 5 | 58 |
| GO:0005840 | ribosome | 6.99e-02 | 1.00e+00 | 3.790 | 1 | 2 | 59 |
| GO:0045216 | cell-cell junction organization | 6.99e-02 | 1.00e+00 | 3.790 | 1 | 2 | 59 |
| GO:0008013 | beta-catenin binding | 7.11e-02 | 1.00e+00 | 3.766 | 1 | 4 | 60 |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 7.22e-02 | 1.00e+00 | 3.742 | 1 | 3 | 61 |
| GO:0000776 | kinetochore | 7.45e-02 | 1.00e+00 | 3.695 | 1 | 4 | 63 |
| GO:0001558 | regulation of cell growth | 7.79e-02 | 1.00e+00 | 3.628 | 1 | 4 | 66 |
| GO:0071260 | cellular response to mechanical stimulus | 7.79e-02 | 1.00e+00 | 3.628 | 1 | 4 | 66 |
| GO:0005524 | ATP binding | 7.85e-02 | 1.00e+00 | 1.270 | 4 | 46 | 1354 |
| GO:0018105 | peptidyl-serine phosphorylation | 8.13e-02 | 1.00e+00 | 3.564 | 1 | 1 | 69 |
| GO:0050790 | regulation of catalytic activity | 8.13e-02 | 1.00e+00 | 3.564 | 1 | 3 | 69 |
| GO:0034329 | cell junction assembly | 8.35e-02 | 1.00e+00 | 3.523 | 1 | 1 | 71 |
| GO:0060070 | canonical Wnt signaling pathway | 8.81e-02 | 1.00e+00 | 3.444 | 1 | 3 | 75 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 8.92e-02 | 1.00e+00 | 3.425 | 1 | 3 | 76 |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 9.03e-02 | 1.00e+00 | 3.406 | 1 | 5 | 77 |
| GO:0004725 | protein tyrosine phosphatase activity | 9.37e-02 | 1.00e+00 | 3.351 | 1 | 2 | 80 |
| GO:0043197 | dendritic spine | 9.70e-02 | 1.00e+00 | 3.297 | 1 | 2 | 83 |
| GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1.03e-01 | 1.00e+00 | 3.213 | 1 | 3 | 88 |
| GO:0009887 | organ morphogenesis | 1.04e-01 | 1.00e+00 | 3.197 | 1 | 2 | 89 |
| GO:0016363 | nuclear matrix | 1.07e-01 | 1.00e+00 | 3.149 | 1 | 11 | 92 |
| GO:0006928 | cellular component movement | 1.07e-01 | 1.00e+00 | 3.149 | 1 | 7 | 92 |
| GO:0035335 | peptidyl-tyrosine dephosphorylation | 1.09e-01 | 1.00e+00 | 3.118 | 1 | 2 | 94 |
| GO:0001649 | osteoblast differentiation | 1.10e-01 | 1.00e+00 | 3.103 | 1 | 6 | 95 |
| GO:0001764 | neuron migration | 1.11e-01 | 1.00e+00 | 3.088 | 1 | 2 | 96 |
| GO:0006468 | protein phosphorylation | 1.11e-01 | 1.00e+00 | 1.805 | 2 | 10 | 467 |
| GO:0006364 | rRNA processing | 1.11e-01 | 1.00e+00 | 3.088 | 1 | 5 | 96 |
| GO:0005178 | integrin binding | 1.12e-01 | 1.00e+00 | 3.073 | 1 | 2 | 97 |
| GO:0071456 | cellular response to hypoxia | 1.14e-01 | 1.00e+00 | 3.058 | 1 | 4 | 98 |
| GO:0014069 | postsynaptic density | 1.22e-01 | 1.00e+00 | 2.945 | 1 | 1 | 106 |
| GO:0005938 | cell cortex | 1.25e-01 | 1.00e+00 | 2.904 | 1 | 3 | 109 |
| GO:0042127 | regulation of cell proliferation | 1.28e-01 | 1.00e+00 | 2.878 | 1 | 4 | 111 |
| GO:0015630 | microtubule cytoskeleton | 1.29e-01 | 1.00e+00 | 2.865 | 1 | 5 | 112 |
| GO:0048015 | phosphatidylinositol-mediated signaling | 1.32e-01 | 1.00e+00 | 2.827 | 1 | 3 | 115 |
| GO:0005635 | nuclear envelope | 1.33e-01 | 1.00e+00 | 2.815 | 1 | 6 | 116 |
| GO:0048471 | perinuclear region of cytoplasm | 1.33e-01 | 1.00e+00 | 1.642 | 2 | 12 | 523 |
| GO:0072562 | blood microparticle | 1.33e-01 | 1.00e+00 | 2.815 | 1 | 4 | 116 |
| GO:0006325 | chromatin organization | 1.40e-01 | 1.00e+00 | 2.730 | 1 | 4 | 123 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 1.43e-01 | 1.00e+00 | 2.707 | 1 | 4 | 125 |
| GO:0007219 | Notch signaling pathway | 1.43e-01 | 1.00e+00 | 2.707 | 1 | 4 | 125 |
| GO:0008201 | heparin binding | 1.45e-01 | 1.00e+00 | 2.684 | 1 | 2 | 127 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 1.45e-01 | 1.00e+00 | 2.684 | 1 | 5 | 127 |
| GO:0030036 | actin cytoskeleton organization | 1.47e-01 | 1.00e+00 | 2.661 | 1 | 5 | 129 |
| GO:0016477 | cell migration | 1.49e-01 | 1.00e+00 | 2.639 | 1 | 6 | 131 |
| GO:0009615 | response to virus | 1.50e-01 | 1.00e+00 | 2.628 | 1 | 6 | 132 |
| GO:0000790 | nuclear chromatin | 1.51e-01 | 1.00e+00 | 2.617 | 1 | 7 | 133 |
| GO:0006644 | phospholipid metabolic process | 1.55e-01 | 1.00e+00 | 2.574 | 1 | 5 | 137 |
| GO:0016887 | ATPase activity | 1.63e-01 | 1.00e+00 | 2.503 | 1 | 7 | 144 |
| GO:0061024 | membrane organization | 1.65e-01 | 1.00e+00 | 2.483 | 1 | 5 | 146 |
| GO:0006457 | protein folding | 1.68e-01 | 1.00e+00 | 2.453 | 1 | 8 | 149 |
| GO:0008017 | microtubule binding | 1.69e-01 | 1.00e+00 | 2.444 | 1 | 7 | 150 |
| GO:0045087 | innate immune response | 1.73e-01 | 1.00e+00 | 1.406 | 2 | 20 | 616 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.77e-01 | 1.00e+00 | 2.369 | 1 | 8 | 158 |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | 1.78e-01 | 1.00e+00 | 2.360 | 1 | 4 | 159 |
| GO:0045121 | membrane raft | 1.80e-01 | 1.00e+00 | 2.342 | 1 | 8 | 161 |
| GO:0034220 | ion transmembrane transport | 1.86e-01 | 1.00e+00 | 2.289 | 1 | 2 | 167 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.87e-01 | 1.00e+00 | 2.280 | 1 | 5 | 168 |
| GO:0006397 | mRNA processing | 1.88e-01 | 1.00e+00 | 2.272 | 1 | 3 | 169 |
| GO:0038095 | Fc-epsilon receptor signaling pathway | 1.90e-01 | 1.00e+00 | 2.255 | 1 | 7 | 171 |
| GO:0016607 | nuclear speck | 1.94e-01 | 1.00e+00 | 2.221 | 1 | 4 | 175 |
| GO:0031965 | nuclear membrane | 1.95e-01 | 1.00e+00 | 2.213 | 1 | 4 | 176 |
| GO:0003714 | transcription corepressor activity | 1.98e-01 | 1.00e+00 | 2.189 | 1 | 7 | 179 |
| GO:0032403 | protein complex binding | 2.04e-01 | 1.00e+00 | 2.141 | 1 | 7 | 185 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 2.10e-01 | 1.00e+00 | 2.095 | 1 | 4 | 191 |
| GO:0003924 | GTPase activity | 2.22e-01 | 1.00e+00 | 2.007 | 1 | 9 | 203 |
| GO:0006184 | GTP catabolic process | 2.38e-01 | 1.00e+00 | 1.891 | 1 | 9 | 220 |
| GO:0007067 | mitotic nuclear division | 2.48e-01 | 1.00e+00 | 1.821 | 1 | 13 | 231 |
| GO:0008380 | RNA splicing | 2.49e-01 | 1.00e+00 | 1.815 | 1 | 13 | 232 |
| GO:0004842 | ubiquitin-protein transferase activity | 2.71e-01 | 1.00e+00 | 1.673 | 1 | 4 | 256 |
| GO:0006281 | DNA repair | 2.78e-01 | 1.00e+00 | 1.628 | 1 | 22 | 264 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 2.86e-01 | 1.00e+00 | 1.580 | 1 | 6 | 273 |
| GO:0043065 | positive regulation of apoptotic process | 2.87e-01 | 1.00e+00 | 1.574 | 1 | 8 | 274 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 2.88e-01 | 1.00e+00 | 1.569 | 1 | 6 | 275 |
| GO:0042493 | response to drug | 3.00e-01 | 1.00e+00 | 1.503 | 1 | 11 | 288 |
| GO:0016567 | protein ubiquitination | 3.09e-01 | 1.00e+00 | 1.449 | 1 | 5 | 299 |
| GO:0043234 | protein complex | 3.10e-01 | 1.00e+00 | 1.444 | 1 | 17 | 300 |
| GO:0006200 | ATP catabolic process | 3.13e-01 | 1.00e+00 | 1.429 | 1 | 14 | 303 |
| GO:0005856 | cytoskeleton | 3.20e-01 | 1.00e+00 | 1.392 | 1 | 8 | 311 |
| GO:0035556 | intracellular signal transduction | 3.25e-01 | 1.00e+00 | 1.364 | 1 | 6 | 317 |
| GO:0030154 | cell differentiation | 3.31e-01 | 1.00e+00 | 1.328 | 1 | 5 | 325 |
| GO:0005525 | GTP binding | 3.34e-01 | 1.00e+00 | 1.315 | 1 | 11 | 328 |
| GO:0007268 | synaptic transmission | 3.54e-01 | 1.00e+00 | 1.209 | 1 | 2 | 353 |
| GO:0005739 | mitochondrion | 3.70e-01 | 1.00e+00 | 0.642 | 2 | 24 | 1046 |
| GO:0007155 | cell adhesion | 3.79e-01 | 1.00e+00 | 1.088 | 1 | 8 | 384 |
| GO:0006508 | proteolysis | 3.99e-01 | 1.00e+00 | 0.993 | 1 | 9 | 410 |
| GO:0007596 | blood coagulation | 4.39e-01 | 1.00e+00 | 0.815 | 1 | 14 | 464 |
| GO:0005886 | plasma membrane | 4.67e-01 | 1.00e+00 | 0.204 | 4 | 38 | 2834 |
| GO:0055085 | transmembrane transport | 4.73e-01 | 1.00e+00 | 0.667 | 1 | 8 | 514 |
| GO:0042803 | protein homodimerization activity | 5.38e-01 | 1.00e+00 | 0.403 | 1 | 11 | 617 |
| GO:0005789 | endoplasmic reticulum membrane | 5.49e-01 | 1.00e+00 | 0.360 | 1 | 10 | 636 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 6.39e-01 | 1.00e+00 | 0.009 | 1 | 19 | 811 |
| GO:0005615 | extracellular space | 7.22e-01 | 1.00e+00 | -0.308 | 1 | 17 | 1010 |
| GO:0008270 | zinc ion binding | 7.42e-01 | 1.00e+00 | -0.387 | 1 | 12 | 1067 |
| GO:0006351 | transcription, DNA-templated | 8.71e-01 | 1.00e+00 | -0.958 | 1 | 25 | 1585 |