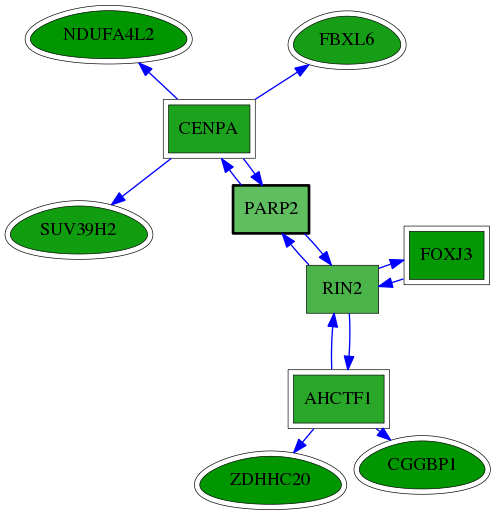
| GO:0000775 | chromosome, centromeric region | 7.58e-04 | 1.00e+00 | 5.574 | 2 | 6 | 41 |
| GO:0000939 | condensed chromosome inner kinetochore | 1.02e-03 | 1.00e+00 | 9.931 | 1 | 1 | 1 |
| GO:0000785 | chromatin | 1.57e-03 | 1.00e+00 | 5.049 | 2 | 6 | 59 |
| GO:0036123 | histone H3-K9 dimethylation | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 3 |
| GO:0000778 | condensed nuclear chromosome kinetochore | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 2 | 3 |
| GO:0071459 | protein localization to chromosome, centromeric region | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 3 |
| GO:0006333 | chromatin assembly or disassembly | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 4 |
| GO:0036124 | histone H3-K9 trimethylation | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 2 | 4 |
| GO:0042754 | negative regulation of circadian rhythm | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 2 | 6 |
| GO:0046974 | histone methyltransferase activity (H3-K9 specific) | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 6 |
| GO:0007140 | male meiosis | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 7 |
| GO:0000780 | condensed nuclear chromosome, centromeric region | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 7 |
| GO:0051382 | kinetochore assembly | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 2 | 7 |
| GO:0051292 | nuclear pore complex assembly | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 4 | 8 |
| GO:0031080 | nuclear pore outer ring | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 2 | 9 |
| GO:0006471 | protein ADP-ribosylation | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 10 |
| GO:0000132 | establishment of mitotic spindle orientation | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 2 | 12 |
| GO:0016409 | palmitoyltransferase activity | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 1 | 12 |
| GO:0003950 | NAD+ ADP-ribosyltransferase activity | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 13 |
| GO:0018345 | protein palmitoylation | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 15 |
| GO:0017112 | Rab guanyl-nucleotide exchange factor activity | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 15 |
| GO:0001227 | RNA polymerase II transcription regulatory region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 1.73e-02 | 1.00e+00 | 5.844 | 1 | 2 | 17 |
| GO:0019706 | protein-cysteine S-palmitoyltransferase activity | 1.73e-02 | 1.00e+00 | 5.844 | 1 | 1 | 17 |
| GO:0005083 | small GTPase regulator activity | 1.73e-02 | 1.00e+00 | 5.844 | 1 | 2 | 17 |
| GO:0005720 | nuclear heterochromatin | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 3 | 20 |
| GO:0034080 | CENP-A containing nucleosome assembly | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 2 | 20 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 2.53e-02 | 1.00e+00 | 5.288 | 1 | 3 | 25 |
| GO:0003682 | chromatin binding | 2.60e-02 | 1.00e+00 | 2.954 | 2 | 15 | 252 |
| GO:0097191 | extrinsic apoptotic signaling pathway | 3.03e-02 | 1.00e+00 | 5.025 | 1 | 1 | 30 |
| GO:0006284 | base-excision repair | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 3 | 32 |
| GO:0000910 | cytokinesis | 3.33e-02 | 1.00e+00 | 4.887 | 1 | 2 | 33 |
| GO:0000786 | nucleosome | 3.43e-02 | 1.00e+00 | 4.844 | 1 | 2 | 34 |
| GO:0032851 | positive regulation of Rab GTPase activity | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 2 | 36 |
| GO:0048511 | rhythmic process | 3.83e-02 | 1.00e+00 | 4.684 | 1 | 5 | 38 |
| GO:0000278 | mitotic cell cycle | 3.91e-02 | 1.00e+00 | 2.637 | 2 | 28 | 314 |
| GO:0050790 | regulation of catalytic activity | 4.52e-02 | 1.00e+00 | 4.440 | 1 | 2 | 45 |
| GO:0051028 | mRNA transport | 4.71e-02 | 1.00e+00 | 4.377 | 1 | 5 | 47 |
| GO:0006334 | nucleosome assembly | 5.10e-02 | 1.00e+00 | 4.259 | 1 | 4 | 51 |
| GO:0005643 | nuclear pore | 5.10e-02 | 1.00e+00 | 4.259 | 1 | 12 | 51 |
| GO:0005654 | nucleoplasm | 5.35e-02 | 1.00e+00 | 1.742 | 3 | 68 | 876 |
| GO:0006338 | chromatin remodeling | 5.49e-02 | 1.00e+00 | 4.150 | 1 | 7 | 55 |
| GO:0000777 | condensed chromosome kinetochore | 5.69e-02 | 1.00e+00 | 4.099 | 1 | 6 | 57 |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 6.27e-02 | 1.00e+00 | 3.954 | 1 | 2 | 63 |
| GO:0003677 | DNA binding | 6.50e-02 | 1.00e+00 | 1.629 | 3 | 52 | 947 |
| GO:0006897 | endocytosis | 6.66e-02 | 1.00e+00 | 3.865 | 1 | 2 | 67 |
| GO:0005096 | GTPase activator activity | 6.75e-02 | 1.00e+00 | 3.844 | 1 | 2 | 68 |
| GO:0016363 | nuclear matrix | 7.23e-02 | 1.00e+00 | 3.742 | 1 | 10 | 73 |
| GO:0071456 | cellular response to hypoxia | 7.52e-02 | 1.00e+00 | 3.684 | 1 | 2 | 76 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 7.64e-02 | 1.00e+00 | 2.099 | 2 | 29 | 456 |
| GO:0003690 | double-stranded DNA binding | 8.19e-02 | 1.00e+00 | 3.556 | 1 | 6 | 83 |
| GO:0006351 | transcription, DNA-templated | 8.87e-02 | 1.00e+00 | 1.445 | 3 | 47 | 1076 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 1.16e-01 | 1.00e+00 | 1.747 | 2 | 22 | 582 |
| GO:0031965 | nuclear membrane | 1.25e-01 | 1.00e+00 | 2.909 | 1 | 8 | 130 |
| GO:0006355 | regulation of transcription, DNA-templated | 1.63e-01 | 1.00e+00 | 1.450 | 2 | 31 | 715 |
| GO:0007264 | small GTPase mediated signal transduction | 1.65e-01 | 1.00e+00 | 2.489 | 1 | 8 | 174 |
| GO:0008270 | zinc ion binding | 1.68e-01 | 1.00e+00 | 1.426 | 2 | 39 | 727 |
| GO:0004842 | ubiquitin-protein transferase activity | 1.69e-01 | 1.00e+00 | 2.448 | 1 | 17 | 179 |
| GO:0005575 | cellular_component | 1.76e-01 | 1.00e+00 | 2.385 | 1 | 4 | 187 |
| GO:0016567 | protein ubiquitination | 1.77e-01 | 1.00e+00 | 2.377 | 1 | 14 | 188 |
| GO:0015031 | protein transport | 1.90e-01 | 1.00e+00 | 2.266 | 1 | 10 | 203 |
| GO:0030154 | cell differentiation | 1.90e-01 | 1.00e+00 | 2.259 | 1 | 5 | 204 |
| GO:0006281 | DNA repair | 1.90e-01 | 1.00e+00 | 2.266 | 1 | 24 | 203 |
| GO:0005634 | nucleus | 2.11e-01 | 1.00e+00 | 0.589 | 5 | 158 | 3246 |
| GO:0006508 | proteolysis | 2.40e-01 | 1.00e+00 | 1.887 | 1 | 11 | 264 |
| GO:0046982 | protein heterodimerization activity | 2.41e-01 | 1.00e+00 | 1.882 | 1 | 11 | 265 |
| GO:0043565 | sequence-specific DNA binding | 2.49e-01 | 1.00e+00 | 1.828 | 1 | 8 | 275 |
| GO:0045892 | negative regulation of transcription, DNA-templated | 2.81e-01 | 1.00e+00 | 1.623 | 1 | 17 | 317 |
| GO:0005730 | nucleolus | 3.60e-01 | 1.00e+00 | 0.682 | 2 | 74 | 1217 |
| GO:0016032 | viral process | 3.61e-01 | 1.00e+00 | 1.190 | 1 | 30 | 428 |
| GO:0005829 | cytosol | 5.71e-01 | 1.00e+00 | 0.128 | 2 | 88 | 1787 |
| GO:0016020 | membrane | 7.33e-01 | 1.00e+00 | -0.306 | 1 | 56 | 1207 |
| GO:0016021 | integral component of membrane | 8.17e-01 | 1.00e+00 | -0.644 | 1 | 19 | 1526 |
| GO:0070062 | extracellular vesicular exosome | 8.41e-01 | 1.00e+00 | -0.749 | 1 | 51 | 1641 |
| GO:0005515 | protein binding | 8.66e-01 | 1.00e+00 | -0.493 | 3 | 198 | 4124 |
| GO:0005886 | plasma membrane | 8.67e-01 | 1.00e+00 | -0.869 | 1 | 46 | 1784 |
| GO:0005737 | cytoplasm | 9.57e-01 | 1.00e+00 | -1.431 | 1 | 127 | 2633 |