int-snw-1457
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| wolf-screen-ratio-mammosphere-adherent | 0.950 | 4.10e-16 | 1.76e-03 | 3.13e-02 |
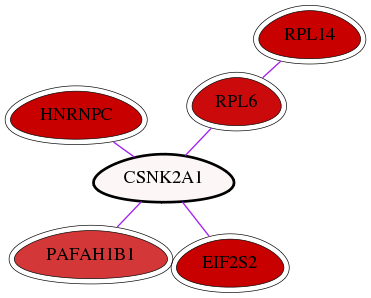
Genes (6)
| Gene Symbol | Entrez Gene ID | Frequency | wolf-screen-ratio-mammosphere-adherent gene score | Best subnetwork score | Degree | wolf adherent-list Hits GI | wolf mammosphere no adherent-list Hits GI |
|---|---|---|---|---|---|---|---|
| [ CSNK2A1 ] | 1457 | 1 | 0.030 | 0.950 | 311 | - | - |
| EIF2S2 | 8894 | 27 | 1.075 | 1.138 | 81 | Yes | - |
| PAFAH1B1 | 5048 | 6 | 0.691 | 1.076 | 71 | Yes | - |
| HNRNPC | 3183 | 20 | 1.812 | 1.026 | 119 | Yes | - |
| RPL14 | 9045 | 42 | 1.250 | 1.113 | 143 | Yes | - |
| RPL6 | 6128 | 11 | 0.844 | 1.113 | 164 | Yes | - |
Interactions (5)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| RPL6 | 6128 | RPL14 | 9045 | pp | -- | int.I2D: BioGrid_Yeast |
| CSNK2A1 | 1457 | PAFAH1B1 | 5048 | pp | -- | int.I2D: HPRD, BCI, BioGrid; int.HPRD: in vitro |
| CSNK2A1 | 1457 | EIF2S2 | 8894 | pp | -- | int.Intact: MI:0217(phosphorylation reaction); int.I2D: BIND, BCI, HPRD; int.HPRD: in vitro |
| CSNK2A1 | 1457 | RPL6 | 6128 | pp | -- | int.Intact: MI:0217(phosphorylation reaction) |
| CSNK2A1 | 1457 | HNRNPC | 3183 | pp | -- | int.I2D: BCI; int.HPRD: in vitro, in vivo |
Related GO terms (137)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0003723 | RNA binding | 4.49e-06 | 6.47e-02 | 4.813 | 4 | 20 | 342 |
| GO:0006413 | translational initiation | 1.44e-05 | 2.07e-01 | 5.783 | 3 | 17 | 131 |
| GO:0005829 | cytosol | 2.67e-05 | 3.86e-01 | 2.531 | 6 | 132 | 2496 |
| GO:0010467 | gene expression | 6.39e-05 | 9.21e-01 | 3.845 | 4 | 59 | 669 |
| GO:0006412 | translation | 7.73e-05 | 1.00e+00 | 4.971 | 3 | 20 | 230 |
| GO:0022625 | cytosolic large ribosomal subunit | 1.68e-04 | 1.00e+00 | 6.616 | 2 | 6 | 49 |
| GO:0044822 | poly(A) RNA binding | 3.80e-04 | 1.00e+00 | 3.187 | 4 | 49 | 1056 |
| GO:0000235 | astral microtubule | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0051660 | establishment of centrosome localization | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0046469 | platelet activating factor metabolic process | 4.16e-04 | 1.00e+00 | 11.231 | 1 | 1 | 1 |
| GO:0019083 | viral transcription | 4.61e-04 | 1.00e+00 | 5.891 | 2 | 10 | 81 |
| GO:0006415 | translational termination | 5.31e-04 | 1.00e+00 | 5.788 | 2 | 10 | 87 |
| GO:0006414 | translational elongation | 6.07e-04 | 1.00e+00 | 5.692 | 2 | 13 | 93 |
| GO:0044267 | cellular protein metabolic process | 6.55e-04 | 1.00e+00 | 3.927 | 3 | 29 | 474 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 7.58e-04 | 1.00e+00 | 5.531 | 2 | 10 | 104 |
| GO:0051081 | nuclear envelope disassembly | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 2 | 2 |
| GO:0002176 | male germ cell proliferation | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0021540 | corpus callosum morphogenesis | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 2 |
| GO:0036035 | osteoclast development | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 2 | 2 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 9.10e-04 | 1.00e+00 | 5.398 | 2 | 11 | 114 |
| GO:0019058 | viral life cycle | 9.26e-04 | 1.00e+00 | 5.386 | 2 | 13 | 115 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0045505 | dynein intermediate chain binding | 1.25e-03 | 1.00e+00 | 9.646 | 1 | 1 | 3 |
| GO:0003735 | structural constituent of ribosome | 1.31e-03 | 1.00e+00 | 5.133 | 2 | 10 | 137 |
| GO:0071174 | mitotic spindle checkpoint | 1.66e-03 | 1.00e+00 | 9.231 | 1 | 1 | 4 |
| GO:0031023 | microtubule organizing center organization | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 5 |
| GO:0008090 | retrograde axon cargo transport | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 2 | 6 |
| GO:0007097 | nuclear migration | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 6 |
| GO:0034452 | dynactin binding | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 6 |
| GO:0001667 | ameboidal-type cell migration | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 6 |
| GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 7 |
| GO:0017145 | stem cell division | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 7 |
| GO:0031512 | motile primary cilium | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 1 | 8 |
| GO:0016071 | mRNA metabolic process | 3.43e-03 | 1.00e+00 | 4.430 | 2 | 31 | 223 |
| GO:0032319 | regulation of Rho GTPase activity | 3.74e-03 | 1.00e+00 | 8.061 | 1 | 2 | 9 |
| GO:0021895 | cerebral cortex neuron differentiation | 3.74e-03 | 1.00e+00 | 8.061 | 1 | 1 | 9 |
| GO:0001675 | acrosome assembly | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 10 |
| GO:0016070 | RNA metabolic process | 4.19e-03 | 1.00e+00 | 4.283 | 2 | 32 | 247 |
| GO:0045502 | dynein binding | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 2 | 11 |
| GO:0045120 | pronucleus | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 2 | 11 |
| GO:0021819 | layer formation in cerebral cortex | 4.57e-03 | 1.00e+00 | 7.772 | 1 | 1 | 11 |
| GO:0070935 | 3'-UTR-mediated mRNA stabilization | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 2 | 12 |
| GO:0016580 | Sin3 complex | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 12 |
| GO:0047496 | vesicle transport along microtubule | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 2 | 12 |
| GO:0042273 | ribosomal large subunit biogenesis | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 4 | 13 |
| GO:0008266 | poly(U) RNA binding | 5.40e-03 | 1.00e+00 | 7.531 | 1 | 1 | 13 |
| GO:0019226 | transmission of nerve impulse | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 14 |
| GO:0048854 | brain morphogenesis | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 15 |
| GO:0016581 | NuRD complex | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0007405 | neuroblast proliferation | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0043274 | phospholipase binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0000132 | establishment of mitotic spindle orientation | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 16 |
| GO:0051879 | Hsp90 protein binding | 7.05e-03 | 1.00e+00 | 7.144 | 1 | 1 | 17 |
| GO:0045773 | positive regulation of axon extension | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 4 | 18 |
| GO:0009306 | protein secretion | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 20 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 9.12e-03 | 1.00e+00 | 6.772 | 1 | 4 | 22 |
| GO:0046329 | negative regulation of JNK cascade | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 1 | 23 |
| GO:0043044 | ATP-dependent chromatin remodeling | 9.53e-03 | 1.00e+00 | 6.708 | 1 | 4 | 23 |
| GO:0008135 | translation factor activity, nucleic acid binding | 9.94e-03 | 1.00e+00 | 6.646 | 1 | 7 | 24 |
| GO:0000278 | mitotic cell cycle | 1.02e-02 | 1.00e+00 | 3.620 | 2 | 48 | 391 |
| GO:0031519 | PcG protein complex | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 4 | 25 |
| GO:0045931 | positive regulation of mitotic cell cycle | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 1 | 26 |
| GO:0003730 | mRNA 3'-UTR binding | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 2 | 26 |
| GO:0007017 | microtubule-based process | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 3 | 27 |
| GO:0030177 | positive regulation of Wnt signaling pathway | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 3 | 27 |
| GO:0031492 | nucleosomal DNA binding | 1.12e-02 | 1.00e+00 | 6.476 | 1 | 4 | 27 |
| GO:0031252 | cell leading edge | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 4 | 28 |
| GO:0010977 | negative regulation of neuron projection development | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 2 | 28 |
| GO:0005875 | microtubule associated complex | 1.16e-02 | 1.00e+00 | 6.424 | 1 | 2 | 28 |
| GO:0061077 | chaperone-mediated protein folding | 1.28e-02 | 1.00e+00 | 6.277 | 1 | 2 | 31 |
| GO:0051219 | phosphoprotein binding | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 2 | 32 |
| GO:0007611 | learning or memory | 1.41e-02 | 1.00e+00 | 6.144 | 1 | 2 | 34 |
| GO:0021766 | hippocampus development | 1.57e-02 | 1.00e+00 | 5.983 | 1 | 4 | 38 |
| GO:0050885 | neuromuscular process controlling balance | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 40 |
| GO:0021987 | cerebral cortex development | 1.73e-02 | 1.00e+00 | 5.839 | 1 | 3 | 42 |
| GO:0008344 | adult locomotory behavior | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 1 | 44 |
| GO:0005871 | kinesin complex | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 3 | 44 |
| GO:0016032 | viral process | 1.86e-02 | 1.00e+00 | 3.170 | 2 | 55 | 534 |
| GO:0048511 | rhythmic process | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 3 | 45 |
| GO:0016042 | lipid catabolic process | 1.98e-02 | 1.00e+00 | 5.646 | 1 | 2 | 48 |
| GO:0003743 | translation initiation factor activity | 2.02e-02 | 1.00e+00 | 5.616 | 1 | 8 | 49 |
| GO:0045732 | positive regulation of protein catabolic process | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 3 | 50 |
| GO:0000226 | microtubule cytoskeleton organization | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 3 | 52 |
| GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding | 2.19e-02 | 1.00e+00 | 5.503 | 1 | 5 | 53 |
| GO:0016020 | membrane | 2.41e-02 | 1.00e+00 | 2.101 | 3 | 90 | 1681 |
| GO:0000776 | kinetochore | 2.59e-02 | 1.00e+00 | 5.254 | 1 | 4 | 63 |
| GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2.59e-02 | 1.00e+00 | 5.254 | 1 | 1 | 63 |
| GO:0030307 | positive regulation of cell growth | 2.84e-02 | 1.00e+00 | 5.123 | 1 | 3 | 69 |
| GO:0008584 | male gonad development | 3.08e-02 | 1.00e+00 | 5.002 | 1 | 3 | 75 |
| GO:0071013 | catalytic step 2 spliceosome | 3.20e-02 | 1.00e+00 | 4.946 | 1 | 4 | 78 |
| GO:0005681 | spliceosomal complex | 3.40e-02 | 1.00e+00 | 4.856 | 1 | 4 | 83 |
| GO:0047485 | protein N-terminus binding | 3.53e-02 | 1.00e+00 | 4.805 | 1 | 5 | 86 |
| GO:0001649 | osteoblast differentiation | 3.77e-02 | 1.00e+00 | 4.708 | 1 | 6 | 92 |
| GO:0001764 | neuron migration | 3.81e-02 | 1.00e+00 | 4.692 | 1 | 3 | 93 |
| GO:0006364 | rRNA processing | 3.85e-02 | 1.00e+00 | 4.677 | 1 | 6 | 94 |
| GO:0030426 | growth cone | 3.93e-02 | 1.00e+00 | 4.646 | 1 | 5 | 96 |
| GO:0005938 | cell cortex | 4.25e-02 | 1.00e+00 | 4.531 | 1 | 3 | 104 |
| GO:0005635 | nuclear envelope | 4.61e-02 | 1.00e+00 | 4.411 | 1 | 6 | 113 |
| GO:0008201 | heparin binding | 4.97e-02 | 1.00e+00 | 4.300 | 1 | 4 | 122 |
| GO:0005515 | protein binding | 4.97e-02 | 1.00e+00 | 0.997 | 5 | 184 | 6024 |
| GO:0030036 | actin cytoskeleton organization | 5.01e-02 | 1.00e+00 | 4.289 | 1 | 5 | 123 |
| GO:0000790 | nuclear chromatin | 5.25e-02 | 1.00e+00 | 4.220 | 1 | 9 | 129 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 5.53e-02 | 1.00e+00 | 4.144 | 1 | 9 | 136 |
| GO:0016055 | Wnt signaling pathway | 5.61e-02 | 1.00e+00 | 4.123 | 1 | 7 | 138 |
| GO:0006355 | regulation of transcription, DNA-templated | 5.94e-02 | 1.00e+00 | 2.270 | 2 | 18 | 997 |
| GO:0008017 | microtubule binding | 5.96e-02 | 1.00e+00 | 4.031 | 1 | 6 | 147 |
| GO:0070062 | extracellular vesicular exosome | 6.20e-02 | 1.00e+00 | 1.587 | 3 | 104 | 2400 |
| GO:0000398 | mRNA splicing, via spliceosome | 6.63e-02 | 1.00e+00 | 3.874 | 1 | 8 | 164 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 6.67e-02 | 1.00e+00 | 3.865 | 1 | 7 | 165 |
| GO:0030424 | axon | 6.71e-02 | 1.00e+00 | 3.856 | 1 | 4 | 166 |
| GO:0031965 | nuclear membrane | 6.79e-02 | 1.00e+00 | 3.839 | 1 | 3 | 168 |
| GO:0001701 | in utero embryonic development | 8.08e-02 | 1.00e+00 | 3.580 | 1 | 8 | 201 |
| GO:0007067 | mitotic nuclear division | 9.08e-02 | 1.00e+00 | 3.405 | 1 | 14 | 227 |
| GO:0008380 | RNA splicing | 9.12e-02 | 1.00e+00 | 3.398 | 1 | 11 | 228 |
| GO:0043025 | neuronal cell body | 9.77e-02 | 1.00e+00 | 3.294 | 1 | 9 | 245 |
| GO:0000166 | nucleotide binding | 1.03e-01 | 1.00e+00 | 3.214 | 1 | 5 | 259 |
| GO:0043234 | protein complex | 1.17e-01 | 1.00e+00 | 3.027 | 1 | 18 | 295 |
| GO:0004674 | protein serine/threonine kinase activity | 1.21e-01 | 1.00e+00 | 2.964 | 1 | 12 | 308 |
| GO:0007411 | axon guidance | 1.26e-01 | 1.00e+00 | 2.914 | 1 | 13 | 319 |
| GO:0005813 | centrosome | 1.28e-01 | 1.00e+00 | 2.882 | 1 | 14 | 326 |
| GO:0007268 | synaptic transmission | 1.30e-01 | 1.00e+00 | 2.856 | 1 | 6 | 332 |
| GO:0005925 | focal adhesion | 1.43e-01 | 1.00e+00 | 2.715 | 1 | 19 | 366 |
| GO:0008284 | positive regulation of cell proliferation | 1.49e-01 | 1.00e+00 | 2.654 | 1 | 8 | 382 |
| GO:0006468 | protein phosphorylation | 1.77e-01 | 1.00e+00 | 2.386 | 1 | 18 | 460 |
| GO:0042802 | identical protein binding | 1.85e-01 | 1.00e+00 | 2.312 | 1 | 20 | 484 |
| GO:0048471 | perinuclear region of cytoplasm | 1.92e-01 | 1.00e+00 | 2.260 | 1 | 13 | 502 |
| GO:0042803 | protein homodimerization activity | 2.23e-01 | 1.00e+00 | 2.014 | 1 | 12 | 595 |
| GO:0005634 | nucleus | 2.86e-01 | 1.00e+00 | 0.662 | 3 | 136 | 4559 |
| GO:0007165 | signal transduction | 3.23e-01 | 1.00e+00 | 1.406 | 1 | 24 | 907 |
| GO:0005654 | nucleoplasm | 3.74e-01 | 1.00e+00 | 1.152 | 1 | 76 | 1082 |
| GO:0003677 | DNA binding | 4.11e-01 | 1.00e+00 | 0.981 | 1 | 28 | 1218 |
| GO:0005524 | ATP binding | 4.32e-01 | 1.00e+00 | 0.889 | 1 | 60 | 1298 |
| GO:0046872 | metal ion binding | 4.35e-01 | 1.00e+00 | 0.879 | 1 | 25 | 1307 |
| GO:0006351 | transcription, DNA-templated | 4.70e-01 | 1.00e+00 | 0.733 | 1 | 31 | 1446 |
| GO:0005730 | nucleolus | 5.16e-01 | 1.00e+00 | 0.551 | 1 | 69 | 1641 |
| GO:0005886 | plasma membrane | 6.94e-01 | 1.00e+00 | -0.103 | 1 | 45 | 2582 |
| GO:0005737 | cytoplasm | 8.37e-01 | 1.00e+00 | -0.648 | 1 | 110 | 3767 |