| GO:0015935 | small ribosomal subunit | 1.42e-08 | 1.38e-04 | 8.931 | 3 | 4 | 12 |
| GO:0006413 | translational initiation | 4.93e-08 | 4.82e-04 | 6.302 | 4 | 12 | 99 |
| GO:0022627 | cytosolic small ribosomal subunit | 2.61e-07 | 2.54e-03 | 7.610 | 3 | 6 | 30 |
| GO:0019083 | viral transcription | 2.30e-06 | 2.25e-02 | 6.586 | 3 | 7 | 61 |
| GO:0006415 | translational termination | 2.54e-06 | 2.48e-02 | 6.539 | 3 | 7 | 63 |
| GO:0006414 | translational elongation | 3.34e-06 | 3.26e-02 | 6.408 | 3 | 7 | 69 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 4.66e-06 | 4.55e-02 | 6.250 | 3 | 7 | 77 |
| GO:0019058 | viral life cycle | 4.85e-06 | 4.73e-02 | 6.231 | 3 | 8 | 78 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6.07e-06 | 5.92e-02 | 6.124 | 3 | 7 | 84 |
| GO:0042274 | ribosomal small subunit biogenesis | 1.15e-05 | 1.12e-01 | 8.472 | 2 | 3 | 11 |
| GO:0003735 | structural constituent of ribosome | 1.33e-05 | 1.30e-01 | 5.748 | 3 | 8 | 109 |
| GO:0016071 | mRNA metabolic process | 5.05e-05 | 4.93e-01 | 5.107 | 3 | 11 | 170 |
| GO:0006412 | translation | 5.80e-05 | 5.66e-01 | 5.041 | 3 | 13 | 178 |
| GO:0016070 | RNA metabolic process | 6.83e-05 | 6.67e-01 | 4.962 | 3 | 11 | 188 |
| GO:0044822 | poly(A) RNA binding | 2.08e-04 | 1.00e+00 | 3.289 | 4 | 25 | 799 |
| GO:0044267 | cellular protein metabolic process | 4.40e-04 | 1.00e+00 | 4.057 | 3 | 13 | 352 |
| GO:0006364 | rRNA processing | 4.58e-04 | 1.00e+00 | 5.865 | 2 | 5 | 67 |
| GO:0002183 | cytoplasmic translational initiation | 5.12e-04 | 1.00e+00 | 10.931 | 1 | 1 | 1 |
| GO:0022605 | oogenesis stage | 5.12e-04 | 1.00e+00 | 10.931 | 1 | 1 | 1 |
| GO:0016032 | viral process | 7.83e-04 | 1.00e+00 | 3.775 | 3 | 13 | 428 |
| GO:0010467 | gene expression | 1.51e-03 | 1.00e+00 | 3.453 | 3 | 17 | 535 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.59e-03 | 1.00e+00 | 4.966 | 2 | 6 | 125 |
| GO:0019082 | viral protein processing | 2.05e-03 | 1.00e+00 | 8.931 | 1 | 1 | 4 |
| GO:0000028 | ribosomal small subunit assembly | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 3 | 5 |
| GO:0002309 | T cell proliferation involved in immune response | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 5 |
| GO:0019221 | cytokine-mediated signaling pathway | 2.98e-03 | 1.00e+00 | 4.505 | 2 | 4 | 172 |
| GO:0006924 | activation-induced cell death of T cells | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 2 | 6 |
| GO:0075733 | intracellular transport of virus | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 2 | 7 |
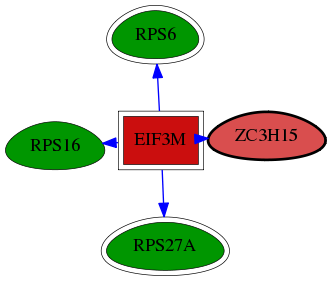
| GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 7 |
| GO:0019068 | virion assembly | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 7 |
| GO:0043065 | positive regulation of apoptotic process | 4.09e-03 | 1.00e+00 | 4.273 | 2 | 3 | 202 |
| GO:0031929 | TOR signaling | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 9 |
| GO:0032479 | regulation of type I interferon production | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 9 |
| GO:0016282 | eukaryotic 43S preinitiation complex | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 1 | 11 |
| GO:0033290 | eukaryotic 48S preinitiation complex | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 1 | 11 |
| GO:0031369 | translation initiation factor binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 13 |
| GO:0048821 | erythrocyte development | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 2 | 13 |
| GO:0005852 | eukaryotic translation initiation factor 3 complex | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 13 |
| GO:0001731 | formation of translation preinitiation complex | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 14 |
| GO:0005978 | glycogen biosynthetic process | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 15 |
| GO:0007220 | Notch receptor processing | 8.17e-03 | 1.00e+00 | 6.931 | 1 | 1 | 16 |
| GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 8.68e-03 | 1.00e+00 | 6.844 | 1 | 1 | 17 |
| GO:0007369 | gastrulation | 8.68e-03 | 1.00e+00 | 6.844 | 1 | 1 | 17 |
| GO:0005844 | polysome | 9.69e-03 | 1.00e+00 | 6.684 | 1 | 1 | 19 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 9.69e-03 | 1.00e+00 | 6.684 | 1 | 4 | 19 |
| GO:0043066 | negative regulation of apoptotic process | 1.01e-02 | 1.00e+00 | 3.601 | 2 | 6 | 322 |
| GO:0032480 | negative regulation of type I interferon production | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 20 |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1.12e-02 | 1.00e+00 | 6.472 | 1 | 1 | 22 |
| GO:0007093 | mitotic cell cycle checkpoint | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 2 | 25 |
| GO:0001890 | placenta development | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 26 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 2 | 26 |
| GO:0033077 | T cell differentiation in thymus | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 26 |
| GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 2 | 28 |
| GO:0016197 | endosomal transport | 1.43e-02 | 1.00e+00 | 6.124 | 1 | 1 | 28 |
| GO:0006446 | regulation of translational initiation | 1.48e-02 | 1.00e+00 | 6.073 | 1 | 2 | 29 |
| GO:0016020 | membrane | 1.55e-02 | 1.00e+00 | 2.279 | 3 | 25 | 1207 |
| GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 1.58e-02 | 1.00e+00 | 5.977 | 1 | 1 | 31 |
| GO:0007254 | JNK cascade | 1.73e-02 | 1.00e+00 | 5.844 | 1 | 1 | 34 |
| GO:0044297 | cell body | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 35 |
| GO:0030666 | endocytic vesicle membrane | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 1 | 36 |
| GO:0003743 | translation initiation factor activity | 1.88e-02 | 1.00e+00 | 5.722 | 1 | 5 | 37 |
| GO:0051403 | stress-activated MAPK cascade | 2.13e-02 | 1.00e+00 | 5.539 | 1 | 2 | 42 |
| GO:0032481 | positive regulation of type I interferon production | 2.43e-02 | 1.00e+00 | 5.347 | 1 | 1 | 48 |
| GO:0034146 | toll-like receptor 5 signaling pathway | 2.53e-02 | 1.00e+00 | 5.288 | 1 | 2 | 50 |
| GO:0034166 | toll-like receptor 10 signaling pathway | 2.53e-02 | 1.00e+00 | 5.288 | 1 | 2 | 50 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 3 | 52 |
| GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2.69e-02 | 1.00e+00 | 5.204 | 1 | 1 | 53 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 2.69e-02 | 1.00e+00 | 5.204 | 1 | 3 | 53 |
| GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway | 2.79e-02 | 1.00e+00 | 5.150 | 1 | 2 | 55 |
| GO:0034162 | toll-like receptor 9 signaling pathway | 2.79e-02 | 1.00e+00 | 5.150 | 1 | 2 | 55 |
| GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway | 2.79e-02 | 1.00e+00 | 5.150 | 1 | 2 | 55 |
| GO:0034134 | toll-like receptor 2 signaling pathway | 2.84e-02 | 1.00e+00 | 5.124 | 1 | 2 | 56 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2.84e-02 | 1.00e+00 | 5.124 | 1 | 3 | 56 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2.89e-02 | 1.00e+00 | 5.099 | 1 | 3 | 57 |
| GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 2.89e-02 | 1.00e+00 | 5.099 | 1 | 2 | 57 |
| GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 2.94e-02 | 1.00e+00 | 5.073 | 1 | 2 | 58 |
| GO:0034138 | toll-like receptor 3 signaling pathway | 2.99e-02 | 1.00e+00 | 5.049 | 1 | 2 | 59 |
| GO:0042593 | glucose homeostasis | 3.04e-02 | 1.00e+00 | 5.025 | 1 | 1 | 60 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 3.04e-02 | 1.00e+00 | 5.025 | 1 | 3 | 60 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 3.09e-02 | 1.00e+00 | 5.001 | 1 | 2 | 61 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 3.09e-02 | 1.00e+00 | 5.001 | 1 | 3 | 61 |
| GO:0000187 | activation of MAPK activity | 3.19e-02 | 1.00e+00 | 4.954 | 1 | 2 | 63 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 3.28e-02 | 1.00e+00 | 4.909 | 1 | 3 | 65 |
| GO:0050852 | T cell receptor signaling pathway | 3.43e-02 | 1.00e+00 | 4.844 | 1 | 1 | 68 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 4 | 70 |
| GO:0034142 | toll-like receptor 4 signaling pathway | 3.68e-02 | 1.00e+00 | 4.742 | 1 | 2 | 73 |
| GO:0071456 | cellular response to hypoxia | 3.83e-02 | 1.00e+00 | 4.684 | 1 | 1 | 76 |
| GO:0006006 | glucose metabolic process | 4.03e-02 | 1.00e+00 | 4.610 | 1 | 2 | 80 |
| GO:0030529 | ribonucleoprotein complex | 4.13e-02 | 1.00e+00 | 4.574 | 1 | 3 | 82 |
| GO:0097190 | apoptotic signaling pathway | 4.18e-02 | 1.00e+00 | 4.556 | 1 | 1 | 83 |
| GO:0002224 | toll-like receptor signaling pathway | 4.18e-02 | 1.00e+00 | 4.556 | 1 | 2 | 83 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 1 | 89 |
| GO:0061024 | membrane organization | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 3 | 89 |
| GO:0005829 | cytosol | 4.57e-02 | 1.00e+00 | 1.713 | 3 | 33 | 1787 |
| GO:0000209 | protein polyubiquitination | 4.62e-02 | 1.00e+00 | 4.408 | 1 | 3 | 92 |
| GO:0007219 | Notch signaling pathway | 4.77e-02 | 1.00e+00 | 4.362 | 1 | 2 | 95 |
| GO:0034220 | ion transmembrane transport | 4.82e-02 | 1.00e+00 | 4.347 | 1 | 1 | 96 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 4.87e-02 | 1.00e+00 | 4.332 | 1 | 3 | 97 |
| GO:0008286 | insulin receptor signaling pathway | 4.87e-02 | 1.00e+00 | 4.332 | 1 | 2 | 97 |
| GO:0010008 | endosome membrane | 5.12e-02 | 1.00e+00 | 4.259 | 1 | 1 | 102 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 5.36e-02 | 1.00e+00 | 4.190 | 1 | 2 | 107 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 5.41e-02 | 1.00e+00 | 4.177 | 1 | 1 | 108 |
| GO:0042981 | regulation of apoptotic process | 5.61e-02 | 1.00e+00 | 4.124 | 1 | 4 | 112 |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | 5.70e-02 | 1.00e+00 | 4.099 | 1 | 1 | 114 |
| GO:0038095 | Fc-epsilon receptor signaling pathway | 6.58e-02 | 1.00e+00 | 3.887 | 1 | 1 | 132 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 7.06e-02 | 1.00e+00 | 3.782 | 1 | 3 | 142 |
| GO:0046872 | metal ion binding | 7.52e-02 | 1.00e+00 | 2.064 | 2 | 10 | 934 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 7.98e-02 | 1.00e+00 | 3.601 | 1 | 3 | 161 |
| GO:0007067 | mitotic nuclear division | 8.17e-02 | 1.00e+00 | 3.565 | 1 | 5 | 165 |
| GO:0030425 | dendrite | 8.22e-02 | 1.00e+00 | 3.556 | 1 | 3 | 166 |
| GO:0005975 | carbohydrate metabolic process | 9.17e-02 | 1.00e+00 | 3.392 | 1 | 1 | 186 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 9.78e-02 | 1.00e+00 | 3.295 | 1 | 2 | 199 |
| GO:0006281 | DNA repair | 9.97e-02 | 1.00e+00 | 3.266 | 1 | 2 | 203 |
| GO:0005515 | protein binding | 1.05e-01 | 1.00e+00 | 0.922 | 4 | 50 | 4124 |
| GO:0019901 | protein kinase binding | 1.13e-01 | 1.00e+00 | 3.073 | 1 | 8 | 232 |
| GO:0005730 | nucleolus | 1.20e-01 | 1.00e+00 | 1.682 | 2 | 21 | 1217 |
| GO:0003723 | RNA binding | 1.20e-01 | 1.00e+00 | 2.983 | 1 | 6 | 247 |
| GO:0005925 | focal adhesion | 1.36e-01 | 1.00e+00 | 2.792 | 1 | 12 | 282 |
| GO:0000278 | mitotic cell cycle | 1.51e-01 | 1.00e+00 | 2.637 | 1 | 9 | 314 |
| GO:0055085 | transmembrane transport | 1.58e-01 | 1.00e+00 | 2.570 | 1 | 1 | 329 |
| GO:0048471 | perinuclear region of cytoplasm | 1.64e-01 | 1.00e+00 | 2.505 | 1 | 7 | 344 |
| GO:0006915 | apoptotic process | 1.93e-01 | 1.00e+00 | 2.248 | 1 | 4 | 411 |
| GO:0070062 | extracellular vesicular exosome | 1.99e-01 | 1.00e+00 | 1.251 | 2 | 28 | 1641 |
| GO:0045087 | innate immune response | 2.11e-01 | 1.00e+00 | 2.111 | 1 | 6 | 452 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.13e-01 | 1.00e+00 | 2.099 | 1 | 6 | 456 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 2.89e-01 | 1.00e+00 | 1.601 | 1 | 8 | 644 |
| GO:0044281 | small molecule metabolic process | 3.64e-01 | 1.00e+00 | 1.210 | 1 | 8 | 844 |
| GO:0005654 | nucleoplasm | 3.75e-01 | 1.00e+00 | 1.157 | 1 | 16 | 876 |
| GO:0005737 | cytoplasm | 4.09e-01 | 1.00e+00 | 0.569 | 2 | 39 | 2633 |
| GO:0006351 | transcription, DNA-templated | 4.42e-01 | 1.00e+00 | 0.860 | 1 | 15 | 1076 |
| GO:0005634 | nucleus | 5.37e-01 | 1.00e+00 | 0.267 | 2 | 40 | 3246 |
| GO:0005886 | plasma membrane | 6.35e-01 | 1.00e+00 | 0.131 | 1 | 13 | 1784 |