| GO:0071987 | WD40-repeat domain binding | 5.12e-04 | 1.00e+00 | 10.931 | 1 | 1 | 1 |
| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1.02e-03 | 1.00e+00 | 9.931 | 1 | 1 | 2 |
| GO:1990077 | primosome complex | 1.02e-03 | 1.00e+00 | 9.931 | 1 | 1 | 2 |
| GO:0072422 | signal transduction involved in DNA damage checkpoint | 1.02e-03 | 1.00e+00 | 9.931 | 1 | 1 | 2 |
| GO:0003896 | DNA primase activity | 1.54e-03 | 1.00e+00 | 9.347 | 1 | 1 | 3 |
| GO:0006269 | DNA replication, synthesis of RNA primer | 1.54e-03 | 1.00e+00 | 9.347 | 1 | 1 | 3 |
| GO:0050658 | RNA transport | 2.05e-03 | 1.00e+00 | 8.931 | 1 | 1 | 4 |
| GO:0030157 | pancreatic juice secretion | 2.05e-03 | 1.00e+00 | 8.931 | 1 | 1 | 4 |
| GO:0000974 | Prp19 complex | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 5 |
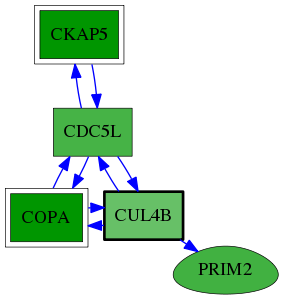
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 5 |
| GO:0000930 | gamma-tubulin complex | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 5 |
| GO:0070914 | UV-damage excision repair | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 6 |
| GO:0030126 | COPI vesicle coat | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 7 |
| GO:0035518 | histone H2A monoubiquitination | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 7 |
| GO:0005662 | DNA replication factor A complex | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 8 |
| GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 4.09e-03 | 1.00e+00 | 7.931 | 1 | 1 | 8 |
| GO:0007051 | spindle organization | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 10 |
| GO:0051297 | centrosome organization | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 12 |
| GO:0035371 | microtubule plus-end | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 13 |
| GO:0006270 | DNA replication initiation | 8.68e-03 | 1.00e+00 | 6.844 | 1 | 1 | 17 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 8.68e-03 | 1.00e+00 | 6.844 | 1 | 1 | 17 |
| GO:0000278 | mitotic cell cycle | 9.66e-03 | 1.00e+00 | 3.637 | 2 | 9 | 314 |
| GO:0032201 | telomere maintenance via semi-conservative replication | 1.02e-02 | 1.00e+00 | 6.610 | 1 | 1 | 20 |
| GO:0051539 | 4 iron, 4 sulfur cluster binding | 1.07e-02 | 1.00e+00 | 6.539 | 1 | 1 | 21 |
| GO:0000722 | telomere maintenance via recombination | 1.12e-02 | 1.00e+00 | 6.472 | 1 | 1 | 22 |
| GO:0006271 | DNA strand elongation involved in DNA replication | 1.48e-02 | 1.00e+00 | 6.073 | 1 | 1 | 29 |
| GO:0016020 | membrane | 1.55e-02 | 1.00e+00 | 2.279 | 3 | 25 | 1207 |
| GO:0045732 | positive regulation of protein catabolic process | 1.73e-02 | 1.00e+00 | 5.844 | 1 | 1 | 34 |
| GO:0003684 | damaged DNA binding | 2.08e-02 | 1.00e+00 | 5.574 | 1 | 1 | 41 |
| GO:0000723 | telomere maintenance | 2.38e-02 | 1.00e+00 | 5.377 | 1 | 1 | 47 |
| GO:0005179 | hormone activity | 3.04e-02 | 1.00e+00 | 5.025 | 1 | 1 | 60 |
| GO:0071013 | catalytic step 2 spliceosome | 3.09e-02 | 1.00e+00 | 5.001 | 1 | 1 | 61 |
| GO:0000922 | spindle pole | 3.43e-02 | 1.00e+00 | 4.844 | 1 | 3 | 68 |
| GO:0015630 | microtubule cytoskeleton | 3.78e-02 | 1.00e+00 | 4.703 | 1 | 2 | 75 |
| GO:0006886 | intracellular protein transport | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 1 | 89 |
| GO:0061024 | membrane organization | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 3 | 89 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 4.62e-02 | 1.00e+00 | 4.408 | 1 | 1 | 92 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 4.87e-02 | 1.00e+00 | 4.332 | 1 | 3 | 97 |
| GO:0005198 | structural molecule activity | 5.07e-02 | 1.00e+00 | 4.273 | 1 | 1 | 101 |
| GO:0007049 | cell cycle | 5.75e-02 | 1.00e+00 | 4.086 | 1 | 2 | 115 |
| GO:0016607 | nuclear speck | 6.09e-02 | 1.00e+00 | 4.001 | 1 | 2 | 122 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 6.24e-02 | 1.00e+00 | 3.966 | 1 | 6 | 125 |
| GO:0000398 | mRNA splicing, via spliceosome | 6.39e-02 | 1.00e+00 | 3.931 | 1 | 2 | 128 |
| GO:0031625 | ubiquitin protein ligase binding | 6.87e-02 | 1.00e+00 | 3.823 | 1 | 2 | 138 |
| GO:0003677 | DNA binding | 7.71e-02 | 1.00e+00 | 2.044 | 2 | 12 | 947 |
| GO:0007067 | mitotic nuclear division | 8.17e-02 | 1.00e+00 | 3.565 | 1 | 5 | 165 |
| GO:0043234 | protein complex | 1.03e-01 | 1.00e+00 | 3.217 | 1 | 8 | 210 |
| GO:0005813 | centrosome | 1.06e-01 | 1.00e+00 | 3.170 | 1 | 6 | 217 |
| GO:0005730 | nucleolus | 1.20e-01 | 1.00e+00 | 1.682 | 2 | 21 | 1217 |
| GO:0003682 | chromatin binding | 1.23e-01 | 1.00e+00 | 2.954 | 1 | 3 | 252 |
| GO:0005737 | cytoplasm | 1.25e-01 | 1.00e+00 | 1.154 | 3 | 39 | 2633 |
| GO:0070062 | extracellular vesicular exosome | 1.99e-01 | 1.00e+00 | 1.251 | 2 | 28 | 1641 |
| GO:0005829 | cytosol | 2.28e-01 | 1.00e+00 | 1.128 | 2 | 33 | 1787 |
| GO:0005615 | extracellular space | 2.92e-01 | 1.00e+00 | 1.585 | 1 | 2 | 651 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.16e-01 | 1.00e+00 | 1.450 | 1 | 9 | 715 |
| GO:0044822 | poly(A) RNA binding | 3.47e-01 | 1.00e+00 | 1.289 | 1 | 25 | 799 |
| GO:0005515 | protein binding | 3.57e-01 | 1.00e+00 | 0.507 | 3 | 50 | 4124 |
| GO:0005654 | nucleoplasm | 3.75e-01 | 1.00e+00 | 1.157 | 1 | 16 | 876 |
| GO:0046872 | metal ion binding | 3.95e-01 | 1.00e+00 | 1.064 | 1 | 10 | 934 |
| GO:0006351 | transcription, DNA-templated | 4.42e-01 | 1.00e+00 | 0.860 | 1 | 15 | 1076 |
| GO:0005634 | nucleus | 5.37e-01 | 1.00e+00 | 0.267 | 2 | 40 | 3246 |
| GO:0005886 | plasma membrane | 6.35e-01 | 1.00e+00 | 0.131 | 1 | 13 | 1784 |