| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1.02e-03 | 1.00e+00 | 9.931 | 1 | 1 | 2 |
| GO:0044233 | ER-mitochondrion membrane contact site | 1.54e-03 | 1.00e+00 | 9.347 | 1 | 1 | 3 |
| GO:0050658 | RNA transport | 2.05e-03 | 1.00e+00 | 8.931 | 1 | 1 | 4 |
| GO:0030157 | pancreatic juice secretion | 2.05e-03 | 1.00e+00 | 8.931 | 1 | 1 | 4 |
| GO:0000930 | gamma-tubulin complex | 2.56e-03 | 1.00e+00 | 8.610 | 1 | 1 | 5 |
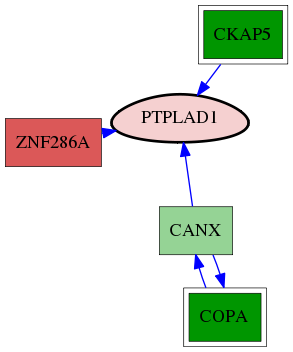
| GO:0030126 | COPI vesicle coat | 3.58e-03 | 1.00e+00 | 8.124 | 1 | 1 | 7 |
| GO:0043234 | protein complex | 4.41e-03 | 1.00e+00 | 4.217 | 2 | 8 | 210 |
| GO:0016601 | Rac protein signal transduction | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 9 |
| GO:0072583 | clathrin-mediated endocytosis | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 9 |
| GO:0007051 | spindle organization | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 10 |
| GO:0048488 | synaptic vesicle endocytosis | 5.11e-03 | 1.00e+00 | 7.610 | 1 | 1 | 10 |
| GO:0009306 | protein secretion | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 2 | 11 |
| GO:0035255 | ionotropic glutamate receptor binding | 5.62e-03 | 1.00e+00 | 7.472 | 1 | 1 | 11 |
| GO:0051297 | centrosome organization | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 12 |
| GO:0016829 | lyase activity | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 12 |
| GO:0035371 | microtubule plus-end | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 13 |
| GO:0034185 | apolipoprotein binding | 6.64e-03 | 1.00e+00 | 7.231 | 1 | 1 | 13 |
| GO:0005790 | smooth endoplasmic reticulum | 7.15e-03 | 1.00e+00 | 7.124 | 1 | 1 | 14 |
| GO:0032839 | dendrite cytoplasm | 7.66e-03 | 1.00e+00 | 7.025 | 1 | 1 | 15 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 8.68e-03 | 1.00e+00 | 6.844 | 1 | 1 | 17 |
| GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane | 9.69e-03 | 1.00e+00 | 6.684 | 1 | 1 | 19 |
| GO:0061077 | chaperone-mediated protein folding | 1.12e-02 | 1.00e+00 | 6.472 | 1 | 1 | 22 |
| GO:0005791 | rough endoplasmic reticulum | 1.17e-02 | 1.00e+00 | 6.408 | 1 | 1 | 23 |
| GO:0007257 | activation of JUN kinase activity | 1.27e-02 | 1.00e+00 | 6.288 | 1 | 1 | 25 |
| GO:0006633 | fatty acid biosynthetic process | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 1 | 26 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 1.32e-02 | 1.00e+00 | 6.231 | 1 | 2 | 26 |
| GO:0005789 | endoplasmic reticulum membrane | 1.45e-02 | 1.00e+00 | 3.335 | 2 | 3 | 387 |
| GO:0016020 | membrane | 1.55e-02 | 1.00e+00 | 2.279 | 3 | 25 | 1207 |
| GO:0007266 | Rho protein signal transduction | 1.78e-02 | 1.00e+00 | 5.802 | 1 | 1 | 35 |
| GO:0001948 | glycoprotein binding | 1.88e-02 | 1.00e+00 | 5.722 | 1 | 1 | 37 |
| GO:0005840 | ribosome | 2.03e-02 | 1.00e+00 | 5.610 | 1 | 3 | 40 |
| GO:0043197 | dendritic spine | 2.58e-02 | 1.00e+00 | 5.259 | 1 | 2 | 51 |
| GO:0005179 | hormone activity | 3.04e-02 | 1.00e+00 | 5.025 | 1 | 1 | 60 |
| GO:0018279 | protein N-linked glycosylation via asparagine | 3.09e-02 | 1.00e+00 | 5.001 | 1 | 1 | 61 |
| GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 3 | 64 |
| GO:0042470 | melanosome | 3.23e-02 | 1.00e+00 | 4.931 | 1 | 2 | 64 |
| GO:0000922 | spindle pole | 3.43e-02 | 1.00e+00 | 4.844 | 1 | 3 | 68 |
| GO:0005096 | GTPase activator activity | 3.43e-02 | 1.00e+00 | 4.844 | 1 | 3 | 68 |
| GO:0051082 | unfolded protein binding | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 1 | 70 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 3.53e-02 | 1.00e+00 | 4.802 | 1 | 4 | 70 |
| GO:0015630 | microtubule cytoskeleton | 3.78e-02 | 1.00e+00 | 4.703 | 1 | 2 | 75 |
| GO:0030246 | carbohydrate binding | 4.03e-02 | 1.00e+00 | 4.610 | 1 | 1 | 80 |
| GO:0006886 | intracellular protein transport | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 1 | 89 |
| GO:0061024 | membrane organization | 4.48e-02 | 1.00e+00 | 4.456 | 1 | 3 | 89 |
| GO:0006457 | protein folding | 4.87e-02 | 1.00e+00 | 4.332 | 1 | 2 | 97 |
| GO:0007568 | aging | 4.87e-02 | 1.00e+00 | 4.332 | 1 | 1 | 97 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 4.87e-02 | 1.00e+00 | 4.332 | 1 | 3 | 97 |
| GO:0005198 | structural molecule activity | 5.07e-02 | 1.00e+00 | 4.273 | 1 | 1 | 101 |
| GO:0005788 | endoplasmic reticulum lumen | 5.21e-02 | 1.00e+00 | 4.231 | 1 | 2 | 104 |
| GO:0043547 | positive regulation of GTPase activity | 5.61e-02 | 1.00e+00 | 4.124 | 1 | 4 | 112 |
| GO:0043687 | post-translational protein modification | 5.80e-02 | 1.00e+00 | 4.073 | 1 | 1 | 116 |
| GO:0030424 | axon | 6.39e-02 | 1.00e+00 | 3.931 | 1 | 4 | 128 |
| GO:0007067 | mitotic nuclear division | 8.17e-02 | 1.00e+00 | 3.565 | 1 | 5 | 165 |
| GO:0043025 | neuronal cell body | 8.46e-02 | 1.00e+00 | 3.514 | 1 | 3 | 171 |
| GO:0007264 | small GTPase mediated signal transduction | 8.60e-02 | 1.00e+00 | 3.489 | 1 | 4 | 174 |
| GO:0005813 | centrosome | 1.06e-01 | 1.00e+00 | 3.170 | 1 | 6 | 217 |
| GO:0005925 | focal adhesion | 1.36e-01 | 1.00e+00 | 2.792 | 1 | 12 | 282 |
| GO:0000278 | mitotic cell cycle | 1.51e-01 | 1.00e+00 | 2.637 | 1 | 9 | 314 |
| GO:0005509 | calcium ion binding | 1.67e-01 | 1.00e+00 | 2.476 | 1 | 3 | 351 |
| GO:0044267 | cellular protein metabolic process | 1.68e-01 | 1.00e+00 | 2.472 | 1 | 13 | 352 |
| GO:0005783 | endoplasmic reticulum | 1.80e-01 | 1.00e+00 | 2.358 | 1 | 3 | 381 |
| GO:0070062 | extracellular vesicular exosome | 1.99e-01 | 1.00e+00 | 1.251 | 2 | 28 | 1641 |
| GO:0005829 | cytosol | 2.28e-01 | 1.00e+00 | 1.128 | 2 | 33 | 1787 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 2.65e-01 | 1.00e+00 | 1.747 | 1 | 9 | 582 |
| GO:0005615 | extracellular space | 2.92e-01 | 1.00e+00 | 1.585 | 1 | 2 | 651 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.16e-01 | 1.00e+00 | 1.450 | 1 | 9 | 715 |
| GO:0044822 | poly(A) RNA binding | 3.47e-01 | 1.00e+00 | 1.289 | 1 | 25 | 799 |
| GO:0005515 | protein binding | 3.57e-01 | 1.00e+00 | 0.507 | 3 | 50 | 4124 |
| GO:0046872 | metal ion binding | 3.95e-01 | 1.00e+00 | 1.064 | 1 | 10 | 934 |
| GO:0003677 | DNA binding | 4.00e-01 | 1.00e+00 | 1.044 | 1 | 12 | 947 |
| GO:0006351 | transcription, DNA-templated | 4.42e-01 | 1.00e+00 | 0.860 | 1 | 15 | 1076 |
| GO:0016021 | integral component of membrane | 5.73e-01 | 1.00e+00 | 0.356 | 1 | 3 | 1526 |
| GO:0005737 | cytoplasm | 7.92e-01 | 1.00e+00 | -0.431 | 1 | 39 | 2633 |
| GO:0005634 | nucleus | 8.67e-01 | 1.00e+00 | -0.733 | 1 | 40 | 3246 |