| GO:0044822 | poly(A) RNA binding | 5.84e-04 | 1.00e+00 | 3.026 | 4 | 25 | 799 |
| GO:0071987 | WD40-repeat domain binding | 6.14e-04 | 1.00e+00 | 10.668 | 1 | 1 | 1 |
| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 2 |
| GO:0072422 | signal transduction involved in DNA damage checkpoint | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 2 |
| GO:0051154 | negative regulation of striated muscle cell differentiation | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 3 |
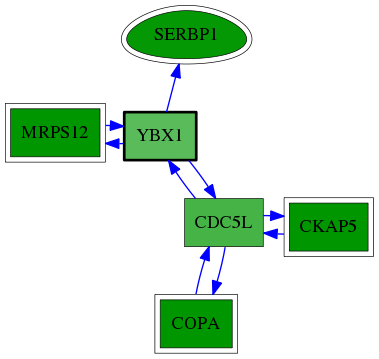
| GO:0050658 | RNA transport | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 4 |
| GO:0030157 | pancreatic juice secretion | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 4 |
| GO:0000398 | mRNA splicing, via spliceosome | 2.47e-03 | 1.00e+00 | 4.668 | 2 | 2 | 128 |
| GO:0016020 | membrane | 2.83e-03 | 1.00e+00 | 2.431 | 4 | 25 | 1207 |
| GO:0000974 | Prp19 complex | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0071204 | histone pre-mRNA 3'end processing complex | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0000930 | gamma-tubulin complex | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0070934 | CRD-mediated mRNA stabilization | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0070937 | CRD-mediated mRNA stability complex | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 6 |
| GO:0030126 | COPI vesicle coat | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 7 |
| GO:0005662 | DNA replication factor A complex | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 8 |
| GO:0007051 | spindle organization | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 10 |
| GO:0043488 | regulation of mRNA stability | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 10 |
| GO:0015935 | small ribosomal subunit | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 4 | 12 |
| GO:0051297 | centrosome organization | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 12 |
| GO:0035371 | microtubule plus-end | 7.96e-03 | 1.00e+00 | 6.968 | 1 | 1 | 13 |
| GO:0005689 | U12-type spliceosomal complex | 1.04e-02 | 1.00e+00 | 6.581 | 1 | 2 | 17 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 1.04e-02 | 1.00e+00 | 6.581 | 1 | 1 | 17 |
| GO:0005761 | mitochondrial ribosome | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 1 | 20 |
| GO:0051781 | positive regulation of cell division | 1.34e-02 | 1.00e+00 | 6.209 | 1 | 1 | 22 |
| GO:0003730 | mRNA 3'-UTR binding | 1.53e-02 | 1.00e+00 | 6.025 | 1 | 1 | 25 |
| GO:0010494 | cytoplasmic stress granule | 1.59e-02 | 1.00e+00 | 5.968 | 1 | 2 | 26 |
| GO:0003697 | single-stranded DNA binding | 3.45e-02 | 1.00e+00 | 4.836 | 1 | 3 | 57 |
| GO:0005179 | hormone activity | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 1 | 60 |
| GO:0071013 | catalytic step 2 spliceosome | 3.69e-02 | 1.00e+00 | 4.738 | 1 | 1 | 61 |
| GO:0000922 | spindle pole | 4.11e-02 | 1.00e+00 | 4.581 | 1 | 3 | 68 |
| GO:0015630 | microtubule cytoskeleton | 4.52e-02 | 1.00e+00 | 4.440 | 1 | 2 | 75 |
| GO:0005737 | cytoplasm | 4.89e-02 | 1.00e+00 | 1.306 | 4 | 39 | 2633 |
| GO:0030529 | ribonucleoprotein complex | 4.94e-02 | 1.00e+00 | 4.311 | 1 | 3 | 82 |
| GO:0003690 | double-stranded DNA binding | 4.99e-02 | 1.00e+00 | 4.293 | 1 | 1 | 83 |
| GO:0005515 | protein binding | 5.22e-02 | 1.00e+00 | 0.981 | 5 | 50 | 4124 |
| GO:0006886 | intracellular protein transport | 5.35e-02 | 1.00e+00 | 4.193 | 1 | 1 | 89 |
| GO:0061024 | membrane organization | 5.35e-02 | 1.00e+00 | 4.193 | 1 | 3 | 89 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 5.82e-02 | 1.00e+00 | 4.069 | 1 | 3 | 97 |
| GO:0005198 | structural molecule activity | 6.05e-02 | 1.00e+00 | 4.010 | 1 | 1 | 101 |
| GO:0070062 | extracellular vesicular exosome | 6.36e-02 | 1.00e+00 | 1.573 | 3 | 28 | 1641 |
| GO:0003735 | structural constituent of ribosome | 6.51e-02 | 1.00e+00 | 3.900 | 1 | 8 | 109 |
| GO:0006355 | regulation of transcription, DNA-templated | 6.59e-02 | 1.00e+00 | 2.187 | 2 | 9 | 715 |
| GO:0042981 | regulation of apoptotic process | 6.69e-02 | 1.00e+00 | 3.861 | 1 | 4 | 112 |
| GO:0016607 | nuclear speck | 7.27e-02 | 1.00e+00 | 3.738 | 1 | 2 | 122 |
| GO:0031965 | nuclear membrane | 7.73e-02 | 1.00e+00 | 3.646 | 1 | 2 | 130 |
| GO:0001701 | in utero embryonic development | 9.10e-02 | 1.00e+00 | 3.402 | 1 | 2 | 154 |
| GO:0007067 | mitotic nuclear division | 9.72e-02 | 1.00e+00 | 3.302 | 1 | 5 | 165 |
| GO:0008380 | RNA splicing | 9.72e-02 | 1.00e+00 | 3.302 | 1 | 4 | 165 |
| GO:0006412 | translation | 1.05e-01 | 1.00e+00 | 3.193 | 1 | 13 | 178 |
| GO:0003677 | DNA binding | 1.08e-01 | 1.00e+00 | 1.781 | 2 | 12 | 947 |
| GO:0043234 | protein complex | 1.22e-01 | 1.00e+00 | 2.954 | 1 | 8 | 210 |
| GO:0005813 | centrosome | 1.26e-01 | 1.00e+00 | 2.907 | 1 | 6 | 217 |
| GO:0043231 | intracellular membrane-bounded organelle | 1.27e-01 | 1.00e+00 | 2.900 | 1 | 1 | 218 |
| GO:0003723 | RNA binding | 1.43e-01 | 1.00e+00 | 2.720 | 1 | 6 | 247 |
| GO:0003682 | chromatin binding | 1.45e-01 | 1.00e+00 | 2.691 | 1 | 3 | 252 |
| GO:0000278 | mitotic cell cycle | 1.78e-01 | 1.00e+00 | 2.374 | 1 | 9 | 314 |
| GO:0006366 | transcription from RNA polymerase II promoter | 1.92e-01 | 1.00e+00 | 2.255 | 1 | 2 | 341 |
| GO:0048471 | perinuclear region of cytoplasm | 1.94e-01 | 1.00e+00 | 2.242 | 1 | 7 | 344 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 2.49e-01 | 1.00e+00 | 1.836 | 1 | 6 | 456 |
| GO:0010467 | gene expression | 2.87e-01 | 1.00e+00 | 1.605 | 1 | 17 | 535 |
| GO:0005829 | cytosol | 3.03e-01 | 1.00e+00 | 0.865 | 2 | 33 | 1787 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 3.08e-01 | 1.00e+00 | 1.484 | 1 | 9 | 582 |
| GO:0005634 | nucleus | 3.18e-01 | 1.00e+00 | 0.589 | 3 | 40 | 3246 |
| GO:0005615 | extracellular space | 3.39e-01 | 1.00e+00 | 1.322 | 1 | 2 | 651 |
| GO:0005654 | nucleoplasm | 4.31e-01 | 1.00e+00 | 0.894 | 1 | 16 | 876 |
| GO:0006351 | transcription, DNA-templated | 5.04e-01 | 1.00e+00 | 0.597 | 1 | 15 | 1076 |
| GO:0005730 | nucleolus | 5.50e-01 | 1.00e+00 | 0.419 | 1 | 21 | 1217 |
| GO:0005886 | plasma membrane | 7.02e-01 | 1.00e+00 | -0.132 | 1 | 13 | 1784 |