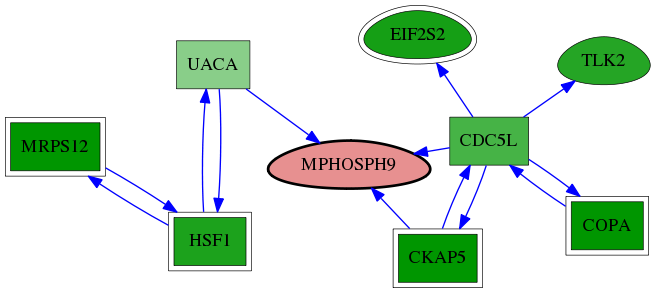
| GO:0071987 | WD40-repeat domain binding | 9.22e-04 | 1.00e+00 | 10.083 | 1 | 1 | 1 |
| GO:0002176 | male germ cell proliferation | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 2 |
| GO:0001672 | regulation of chromatin assembly or disassembly | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 2 |
| GO:0090231 | regulation of spindle checkpoint | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 2 |
| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 2 |
| GO:0072422 | signal transduction involved in DNA damage checkpoint | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 2 |
| GO:0016020 | membrane | 2.35e-03 | 1.00e+00 | 2.168 | 5 | 25 | 1207 |
| GO:0005850 | eukaryotic translation initiation factor 2 complex | 2.76e-03 | 1.00e+00 | 8.499 | 1 | 1 | 3 |
| GO:0043293 | apoptosome | 2.76e-03 | 1.00e+00 | 8.499 | 1 | 1 | 3 |
| GO:0007143 | female meiotic division | 2.76e-03 | 1.00e+00 | 8.499 | 1 | 1 | 3 |
| GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding | 2.76e-03 | 1.00e+00 | 8.499 | 1 | 1 | 3 |
| GO:0050658 | RNA transport | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 4 |
| GO:0030157 | pancreatic juice secretion | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 4 |
| GO:0000974 | Prp19 complex | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 5 |
| GO:0000930 | gamma-tubulin complex | 4.60e-03 | 1.00e+00 | 7.762 | 1 | 1 | 5 |
| GO:0060136 | embryonic process involved in female pregnancy | 5.52e-03 | 1.00e+00 | 7.499 | 1 | 1 | 6 |
| GO:0030126 | COPI vesicle coat | 6.44e-03 | 1.00e+00 | 7.276 | 1 | 1 | 7 |
| GO:0005662 | DNA replication factor A complex | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 8 |
| GO:0042347 | negative regulation of NF-kappaB import into nucleus | 8.27e-03 | 1.00e+00 | 6.914 | 1 | 1 | 9 |
| GO:0071480 | cellular response to gamma radiation | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 1 | 10 |
| GO:0010507 | negative regulation of autophagy | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 1 | 10 |
| GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 1 | 10 |
| GO:0007051 | spindle organization | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 1 | 10 |
| GO:0045120 | pronucleus | 1.01e-02 | 1.00e+00 | 6.624 | 1 | 1 | 11 |
| GO:0006412 | translation | 1.09e-02 | 1.00e+00 | 3.608 | 2 | 13 | 178 |
| GO:0015935 | small ribosomal subunit | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 4 | 12 |
| GO:0042307 | positive regulation of protein import into nucleus | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 2 | 12 |
| GO:0051297 | centrosome organization | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 1 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 1 | 12 |
| GO:0035371 | microtubule plus-end | 1.19e-02 | 1.00e+00 | 6.383 | 1 | 1 | 13 |
| GO:0008135 | translation factor activity, nucleic acid binding | 1.37e-02 | 1.00e+00 | 6.177 | 1 | 4 | 15 |
| GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 1.37e-02 | 1.00e+00 | 6.177 | 1 | 1 | 15 |
| GO:0043234 | protein complex | 1.50e-02 | 1.00e+00 | 3.369 | 2 | 8 | 210 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 1.56e-02 | 1.00e+00 | 5.996 | 1 | 1 | 17 |
| GO:0005813 | centrosome | 1.60e-02 | 1.00e+00 | 3.322 | 2 | 6 | 217 |
| GO:0001892 | embryonic placenta development | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 1 | 20 |
| GO:0032720 | negative regulation of tumor necrosis factor production | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 1 | 20 |
| GO:0005761 | mitochondrial ribosome | 1.83e-02 | 1.00e+00 | 5.762 | 1 | 1 | 20 |
| GO:0034605 | cellular response to heat | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 1 | 22 |
| GO:0040018 | positive regulation of multicellular organism growth | 2.01e-02 | 1.00e+00 | 5.624 | 1 | 1 | 22 |
| GO:0003682 | chromatin binding | 2.12e-02 | 1.00e+00 | 3.106 | 2 | 3 | 252 |
| GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2.91e-02 | 1.00e+00 | 5.083 | 1 | 2 | 32 |
| GO:0006468 | protein phosphorylation | 2.95e-02 | 1.00e+00 | 2.850 | 2 | 3 | 301 |
| GO:0044822 | poly(A) RNA binding | 3.15e-02 | 1.00e+00 | 2.026 | 3 | 25 | 799 |
| GO:0009411 | response to UV | 3.27e-02 | 1.00e+00 | 4.914 | 1 | 1 | 36 |
| GO:0003743 | translation initiation factor activity | 3.36e-02 | 1.00e+00 | 4.874 | 1 | 5 | 37 |
| GO:0006952 | defense response | 3.36e-02 | 1.00e+00 | 4.874 | 1 | 1 | 37 |
| GO:0005515 | protein binding | 3.44e-02 | 1.00e+00 | 0.881 | 7 | 50 | 4124 |
| GO:0005882 | intermediate filament | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 2 | 40 |
| GO:0048471 | perinuclear region of cytoplasm | 3.78e-02 | 1.00e+00 | 2.657 | 2 | 7 | 344 |
| GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 3.81e-02 | 1.00e+00 | 4.691 | 1 | 1 | 42 |
| GO:0005814 | centriole | 4.16e-02 | 1.00e+00 | 4.560 | 1 | 1 | 46 |
| GO:0050728 | negative regulation of inflammatory response | 4.43e-02 | 1.00e+00 | 4.469 | 1 | 1 | 49 |
| GO:0007059 | chromosome segregation | 4.52e-02 | 1.00e+00 | 4.440 | 1 | 1 | 50 |
| GO:0001078 | RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 4.52e-02 | 1.00e+00 | 4.440 | 1 | 1 | 50 |
| GO:0018105 | peptidyl-serine phosphorylation | 4.61e-02 | 1.00e+00 | 4.411 | 1 | 2 | 51 |
| GO:0008584 | male gonad development | 5.13e-02 | 1.00e+00 | 4.251 | 1 | 1 | 57 |
| GO:0005179 | hormone activity | 5.40e-02 | 1.00e+00 | 4.177 | 1 | 1 | 60 |
| GO:0071013 | catalytic step 2 spliceosome | 5.49e-02 | 1.00e+00 | 4.153 | 1 | 1 | 61 |
| GO:0000922 | spindle pole | 6.10e-02 | 1.00e+00 | 3.996 | 1 | 3 | 68 |
| GO:0016568 | chromatin modification | 6.10e-02 | 1.00e+00 | 3.996 | 1 | 2 | 68 |
| GO:0005737 | cytoplasm | 6.58e-02 | 1.00e+00 | 1.043 | 5 | 39 | 2633 |
| GO:0015630 | microtubule cytoskeleton | 6.71e-02 | 1.00e+00 | 3.855 | 1 | 2 | 75 |
| GO:0005635 | nuclear envelope | 7.40e-02 | 1.00e+00 | 3.708 | 1 | 2 | 83 |
| GO:0006886 | intracellular protein transport | 7.91e-02 | 1.00e+00 | 3.608 | 1 | 1 | 89 |
| GO:0061024 | membrane organization | 7.91e-02 | 1.00e+00 | 3.608 | 1 | 3 | 89 |
| GO:0032496 | response to lipopolysaccharide | 8.26e-02 | 1.00e+00 | 3.544 | 1 | 1 | 93 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 8.60e-02 | 1.00e+00 | 3.484 | 1 | 3 | 97 |
| GO:0006413 | translational initiation | 8.77e-02 | 1.00e+00 | 3.454 | 1 | 12 | 99 |
| GO:0005198 | structural molecule activity | 8.94e-02 | 1.00e+00 | 3.425 | 1 | 1 | 101 |
| GO:0005730 | nucleolus | 9.11e-02 | 1.00e+00 | 1.419 | 3 | 21 | 1217 |
| GO:0003735 | structural constituent of ribosome | 9.61e-02 | 1.00e+00 | 3.315 | 1 | 8 | 109 |
| GO:0007049 | cell cycle | 1.01e-01 | 1.00e+00 | 3.238 | 1 | 2 | 115 |
| GO:0006974 | cellular response to DNA damage stimulus | 1.06e-01 | 1.00e+00 | 3.165 | 1 | 2 | 121 |
| GO:0016607 | nuclear speck | 1.07e-01 | 1.00e+00 | 3.153 | 1 | 2 | 122 |
| GO:0000398 | mRNA splicing, via spliceosome | 1.12e-01 | 1.00e+00 | 3.083 | 1 | 2 | 128 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 1.24e-01 | 1.00e+00 | 2.934 | 1 | 4 | 142 |
| GO:0001701 | in utero embryonic development | 1.33e-01 | 1.00e+00 | 2.817 | 1 | 2 | 154 |
| GO:0007067 | mitotic nuclear division | 1.42e-01 | 1.00e+00 | 2.717 | 1 | 5 | 165 |
| GO:0007283 | spermatogenesis | 1.45e-01 | 1.00e+00 | 2.691 | 1 | 2 | 168 |
| GO:0005856 | cytoskeleton | 1.62e-01 | 1.00e+00 | 2.514 | 1 | 3 | 190 |
| GO:0004674 | protein serine/threonine kinase activity | 1.68e-01 | 1.00e+00 | 2.461 | 1 | 3 | 197 |
| GO:0035556 | intracellular signal transduction | 1.69e-01 | 1.00e+00 | 2.447 | 1 | 3 | 199 |
| GO:0003723 | RNA binding | 2.06e-01 | 1.00e+00 | 2.135 | 1 | 6 | 247 |
| GO:0005829 | cytosol | 2.18e-01 | 1.00e+00 | 0.865 | 3 | 33 | 1787 |
| GO:0000139 | Golgi membrane | 2.21e-01 | 1.00e+00 | 2.023 | 1 | 4 | 267 |
| GO:0008150 | biological_process | 2.23e-01 | 1.00e+00 | 2.007 | 1 | 2 | 270 |
| GO:0008285 | negative regulation of cell proliferation | 2.28e-01 | 1.00e+00 | 1.970 | 1 | 3 | 277 |
| GO:0003674 | molecular_function | 2.43e-01 | 1.00e+00 | 1.869 | 1 | 1 | 297 |
| GO:0000278 | mitotic cell cycle | 2.55e-01 | 1.00e+00 | 1.789 | 1 | 9 | 314 |
| GO:0006351 | transcription, DNA-templated | 2.61e-01 | 1.00e+00 | 1.012 | 2 | 15 | 1076 |
| GO:0044267 | cellular protein metabolic process | 2.81e-01 | 1.00e+00 | 1.624 | 1 | 13 | 352 |
| GO:0005794 | Golgi apparatus | 3.24e-01 | 1.00e+00 | 1.387 | 1 | 3 | 415 |
| GO:0005634 | nucleus | 3.47e-01 | 1.00e+00 | 0.419 | 4 | 40 | 3246 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.50e-01 | 1.00e+00 | 1.251 | 1 | 6 | 456 |
| GO:0010467 | gene expression | 3.98e-01 | 1.00e+00 | 1.020 | 1 | 17 | 535 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 4.25e-01 | 1.00e+00 | 0.899 | 1 | 9 | 582 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 4.59e-01 | 1.00e+00 | 0.753 | 1 | 8 | 644 |
| GO:0070062 | extracellular vesicular exosome | 4.62e-01 | 1.00e+00 | 0.403 | 2 | 28 | 1641 |
| GO:0005615 | extracellular space | 4.63e-01 | 1.00e+00 | 0.737 | 1 | 2 | 651 |
| GO:0005739 | mitochondrion | 4.67e-01 | 1.00e+00 | 0.719 | 1 | 4 | 659 |
| GO:0005576 | extracellular region | 4.69e-01 | 1.00e+00 | 0.711 | 1 | 1 | 663 |
| GO:0006355 | regulation of transcription, DNA-templated | 4.96e-01 | 1.00e+00 | 0.602 | 1 | 9 | 715 |
| GO:0005524 | ATP binding | 5.78e-01 | 1.00e+00 | 0.283 | 1 | 10 | 892 |
| GO:0046872 | metal ion binding | 5.96e-01 | 1.00e+00 | 0.216 | 1 | 10 | 934 |
| GO:0003677 | DNA binding | 6.01e-01 | 1.00e+00 | 0.196 | 1 | 12 | 947 |
| GO:0005886 | plasma membrane | 8.37e-01 | 1.00e+00 | -0.717 | 1 | 13 | 1784 |