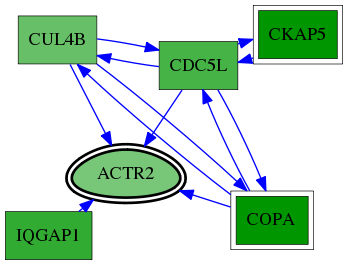
| GO:0071987 | WD40-repeat domain binding | 6.14e-04 | 1.00e+00 | 10.668 | 1 | 1 | 1 |
| GO:0035305 | negative regulation of dephosphorylation | 6.14e-04 | 1.00e+00 | 10.668 | 1 | 1 | 1 |
| GO:0015630 | microtubule cytoskeleton | 8.56e-04 | 1.00e+00 | 5.440 | 2 | 2 | 75 |
| GO:0016344 | meiotic chromosome movement towards spindle pole | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 2 |
| GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 2 |
| GO:0072422 | signal transduction involved in DNA damage checkpoint | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 2 |
| GO:0033206 | meiotic cytokinesis | 1.23e-03 | 1.00e+00 | 9.668 | 1 | 1 | 2 |
| GO:0008356 | asymmetric cell division | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 3 |
| GO:0051653 | spindle localization | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 3 |
| GO:0030478 | actin cap | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 3 |
| GO:0016482 | cytoplasmic transport | 1.84e-03 | 1.00e+00 | 9.083 | 1 | 1 | 3 |
| GO:0050658 | RNA transport | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 4 |
| GO:0030157 | pancreatic juice secretion | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 4 |
| GO:1990138 | neuron projection extension | 2.46e-03 | 1.00e+00 | 8.668 | 1 | 1 | 4 |
| GO:0015629 | actin cytoskeleton | 2.78e-03 | 1.00e+00 | 4.581 | 2 | 3 | 136 |
| GO:0016020 | membrane | 2.83e-03 | 1.00e+00 | 2.431 | 4 | 25 | 1207 |
| GO:0000974 | Prp19 complex | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0072015 | glomerular visceral epithelial cell development | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0000930 | gamma-tubulin complex | 3.07e-03 | 1.00e+00 | 8.347 | 1 | 1 | 5 |
| GO:0070914 | UV-damage excision repair | 3.68e-03 | 1.00e+00 | 8.083 | 1 | 1 | 6 |
| GO:0005885 | Arp2/3 protein complex | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 7 |
| GO:0036057 | slit diaphragm | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 7 |
| GO:0030126 | COPI vesicle coat | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 7 |
| GO:0035518 | histone H2A monoubiquitination | 4.29e-03 | 1.00e+00 | 7.861 | 1 | 1 | 7 |
| GO:0005662 | DNA replication factor A complex | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 8 |
| GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 4.91e-03 | 1.00e+00 | 7.668 | 1 | 1 | 8 |
| GO:0034314 | Arp2/3 complex-mediated actin nucleation | 5.52e-03 | 1.00e+00 | 7.499 | 1 | 1 | 9 |
| GO:0001817 | regulation of cytokine production | 5.52e-03 | 1.00e+00 | 7.499 | 1 | 1 | 9 |
| GO:0005099 | Ras GTPase activator activity | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 10 |
| GO:0007051 | spindle organization | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 10 |
| GO:0005095 | GTPase inhibitor activity | 6.13e-03 | 1.00e+00 | 7.347 | 1 | 1 | 10 |
| GO:0005737 | cytoplasm | 6.61e-03 | 1.00e+00 | 1.628 | 5 | 39 | 2633 |
| GO:0051297 | centrosome organization | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 12 |
| GO:0043539 | protein serine/threonine kinase activator activity | 7.35e-03 | 1.00e+00 | 7.083 | 1 | 1 | 12 |
| GO:0035371 | microtubule plus-end | 7.96e-03 | 1.00e+00 | 6.968 | 1 | 1 | 13 |
| GO:0070062 | extracellular vesicular exosome | 8.95e-03 | 1.00e+00 | 1.988 | 4 | 28 | 1641 |
| GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | 9.18e-03 | 1.00e+00 | 6.762 | 1 | 1 | 15 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 1.04e-02 | 1.00e+00 | 6.581 | 1 | 1 | 17 |
| GO:0071364 | cellular response to epidermal growth factor stimulus | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 1 | 18 |
| GO:0032320 | positive regulation of Ras GTPase activity | 1.10e-02 | 1.00e+00 | 6.499 | 1 | 1 | 18 |
| GO:0005925 | focal adhesion | 1.15e-02 | 1.00e+00 | 3.529 | 2 | 12 | 282 |
| GO:0048365 | Rac GTPase binding | 1.16e-02 | 1.00e+00 | 6.421 | 1 | 1 | 19 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.16e-02 | 1.00e+00 | 6.421 | 1 | 4 | 19 |
| GO:0007163 | establishment or maintenance of cell polarity | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 2 | 20 |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1.22e-02 | 1.00e+00 | 6.347 | 1 | 1 | 20 |
| GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | 1.28e-02 | 1.00e+00 | 6.276 | 1 | 1 | 21 |
| GO:0031252 | cell leading edge | 1.47e-02 | 1.00e+00 | 6.083 | 1 | 2 | 24 |
| GO:0071277 | cellular response to calcium ion | 1.59e-02 | 1.00e+00 | 5.968 | 1 | 1 | 26 |
| GO:0005884 | actin filament | 1.65e-02 | 1.00e+00 | 5.914 | 1 | 1 | 27 |
| GO:0016328 | lateral plasma membrane | 1.89e-02 | 1.00e+00 | 5.714 | 1 | 2 | 31 |
| GO:0045732 | positive regulation of protein catabolic process | 2.07e-02 | 1.00e+00 | 5.581 | 1 | 1 | 34 |
| GO:0045860 | positive regulation of protein kinase activity | 2.25e-02 | 1.00e+00 | 5.459 | 1 | 1 | 37 |
| GO:0042995 | cell projection | 2.49e-02 | 1.00e+00 | 5.311 | 1 | 1 | 41 |
| GO:0003684 | damaged DNA binding | 2.49e-02 | 1.00e+00 | 5.311 | 1 | 1 | 41 |
| GO:0043086 | negative regulation of catalytic activity | 2.73e-02 | 1.00e+00 | 5.177 | 1 | 1 | 45 |
| GO:0019903 | protein phosphatase binding | 2.85e-02 | 1.00e+00 | 5.114 | 1 | 1 | 47 |
| GO:0005730 | nucleolus | 2.89e-02 | 1.00e+00 | 2.004 | 3 | 21 | 1217 |
| GO:0042384 | cilium assembly | 3.21e-02 | 1.00e+00 | 4.941 | 1 | 1 | 53 |
| GO:0050796 | regulation of insulin secretion | 3.27e-02 | 1.00e+00 | 4.914 | 1 | 1 | 54 |
| GO:0005179 | hormone activity | 3.63e-02 | 1.00e+00 | 4.762 | 1 | 1 | 60 |
| GO:0071013 | catalytic step 2 spliceosome | 3.69e-02 | 1.00e+00 | 4.738 | 1 | 1 | 61 |
| GO:0051015 | actin filament binding | 3.81e-02 | 1.00e+00 | 4.691 | 1 | 1 | 63 |
| GO:0030426 | growth cone | 3.87e-02 | 1.00e+00 | 4.668 | 1 | 3 | 64 |
| GO:0006112 | energy reserve metabolic process | 3.93e-02 | 1.00e+00 | 4.646 | 1 | 2 | 65 |
| GO:0000922 | spindle pole | 4.11e-02 | 1.00e+00 | 4.581 | 1 | 3 | 68 |
| GO:0005096 | GTPase activator activity | 4.11e-02 | 1.00e+00 | 4.581 | 1 | 3 | 68 |
| GO:0005200 | structural constituent of cytoskeleton | 4.11e-02 | 1.00e+00 | 4.581 | 1 | 4 | 68 |
| GO:0006928 | cellular component movement | 4.23e-02 | 1.00e+00 | 4.539 | 1 | 3 | 70 |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 4.23e-02 | 1.00e+00 | 4.539 | 1 | 3 | 70 |
| GO:0030496 | midbody | 4.40e-02 | 1.00e+00 | 4.479 | 1 | 3 | 73 |
| GO:0006886 | intracellular protein transport | 5.35e-02 | 1.00e+00 | 4.193 | 1 | 1 | 89 |
| GO:0061024 | membrane organization | 5.35e-02 | 1.00e+00 | 4.193 | 1 | 3 | 89 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 5.52e-02 | 1.00e+00 | 4.145 | 1 | 1 | 92 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 5.82e-02 | 1.00e+00 | 4.069 | 1 | 3 | 97 |
| GO:0005516 | calmodulin binding | 5.93e-02 | 1.00e+00 | 4.039 | 1 | 2 | 99 |
| GO:0005198 | structural molecule activity | 6.05e-02 | 1.00e+00 | 4.010 | 1 | 1 | 101 |
| GO:0043547 | positive regulation of GTPase activity | 6.69e-02 | 1.00e+00 | 3.861 | 1 | 4 | 112 |
| GO:0043005 | neuron projection | 6.75e-02 | 1.00e+00 | 3.848 | 1 | 2 | 113 |
| GO:0007049 | cell cycle | 6.86e-02 | 1.00e+00 | 3.823 | 1 | 2 | 115 |
| GO:0016607 | nuclear speck | 7.27e-02 | 1.00e+00 | 3.738 | 1 | 2 | 122 |
| GO:0030424 | axon | 7.61e-02 | 1.00e+00 | 3.668 | 1 | 4 | 128 |
| GO:0000398 | mRNA splicing, via spliceosome | 7.61e-02 | 1.00e+00 | 3.668 | 1 | 2 | 128 |
| GO:0032403 | protein complex binding | 7.73e-02 | 1.00e+00 | 3.646 | 1 | 2 | 130 |
| GO:0005829 | cytosol | 7.91e-02 | 1.00e+00 | 1.450 | 3 | 33 | 1787 |
| GO:0031625 | ubiquitin protein ligase binding | 8.19e-02 | 1.00e+00 | 3.560 | 1 | 2 | 138 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 8.42e-02 | 1.00e+00 | 3.519 | 1 | 3 | 142 |
| GO:0005874 | microtubule | 9.61e-02 | 1.00e+00 | 3.320 | 1 | 2 | 163 |
| GO:0007067 | mitotic nuclear division | 9.72e-02 | 1.00e+00 | 3.302 | 1 | 5 | 165 |
| GO:0007264 | small GTPase mediated signal transduction | 1.02e-01 | 1.00e+00 | 3.225 | 1 | 4 | 174 |
| GO:0043234 | protein complex | 1.22e-01 | 1.00e+00 | 2.954 | 1 | 8 | 210 |
| GO:0005813 | centrosome | 1.26e-01 | 1.00e+00 | 2.907 | 1 | 6 | 217 |
| GO:0030054 | cell junction | 1.27e-01 | 1.00e+00 | 2.900 | 1 | 3 | 218 |
| GO:0019901 | protein kinase binding | 1.34e-01 | 1.00e+00 | 2.810 | 1 | 8 | 232 |
| GO:0003682 | chromatin binding | 1.45e-01 | 1.00e+00 | 2.691 | 1 | 3 | 252 |
| GO:0000278 | mitotic cell cycle | 1.78e-01 | 1.00e+00 | 2.374 | 1 | 9 | 314 |
| GO:0005509 | calcium ion binding | 1.97e-01 | 1.00e+00 | 2.213 | 1 | 3 | 351 |
| GO:0005515 | protein binding | 2.11e-01 | 1.00e+00 | 0.659 | 4 | 50 | 4124 |
| GO:0045087 | innate immune response | 2.48e-01 | 1.00e+00 | 1.848 | 1 | 6 | 452 |
| GO:0005886 | plasma membrane | 3.02e-01 | 1.00e+00 | 0.868 | 2 | 13 | 1784 |
| GO:0005634 | nucleus | 3.18e-01 | 1.00e+00 | 0.589 | 3 | 40 | 3246 |
| GO:0007165 | signal transduction | 3.25e-01 | 1.00e+00 | 1.397 | 1 | 5 | 618 |
| GO:0005615 | extracellular space | 3.39e-01 | 1.00e+00 | 1.322 | 1 | 2 | 651 |
| GO:0006355 | regulation of transcription, DNA-templated | 3.66e-01 | 1.00e+00 | 1.187 | 1 | 9 | 715 |
| GO:0044822 | poly(A) RNA binding | 4.01e-01 | 1.00e+00 | 1.026 | 1 | 25 | 799 |
| GO:0044281 | small molecule metabolic process | 4.19e-01 | 1.00e+00 | 0.947 | 1 | 8 | 844 |
| GO:0005524 | ATP binding | 4.37e-01 | 1.00e+00 | 0.868 | 1 | 10 | 892 |
| GO:0003677 | DNA binding | 4.58e-01 | 1.00e+00 | 0.781 | 1 | 12 | 947 |
| GO:0006351 | transcription, DNA-templated | 5.04e-01 | 1.00e+00 | 0.597 | 1 | 15 | 1076 |