int-snw-388
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| tai-screen-luciferase | 6.647 | 2.43e-150 | 5.53e-08 | 1.14e-03 |
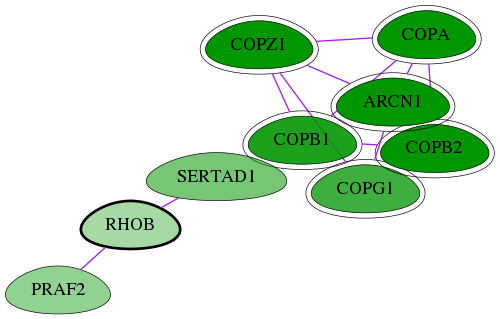
Genes (9)
| Gene Symbol | Entrez Gene ID | Frequency | tai-screen-luciferase gene score | Best subnetwork score | Degree | List-Gonzales GI | Tai-Hits |
|---|---|---|---|---|---|---|---|
| COPZ1 | 22818 | 32 | -8.301 | 9.063 | 13 | Yes | Yes |
| COPA | 1314 | 32 | -9.395 | 9.063 | 170 | Yes | Yes |
| PRAF2 | 11230 | 1 | -3.015 | 6.647 | 6 | - | - |
| SERTAD1 | 29950 | 1 | -3.746 | 6.647 | 37 | - | - |
| ARCN1 | 372 | 32 | -8.232 | 9.063 | 118 | Yes | Yes |
| COPB1 | 1315 | 22 | -6.221 | 9.063 | 118 | Yes | Yes |
| COPG1 | 22820 | 9 | -5.279 | 7.138 | 37 | Yes | Yes |
| COPB2 | 9276 | 32 | -13.168 | 9.063 | 41 | Yes | Yes |
| [ RHOB ] | 388 | 1 | -2.468 | 6.647 | 30 | - | - |
Interactions (16)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| COPA | 1314 | COPB1 | 1315 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct_Yeast, MINT_Yeast, Krogan_Core, YeastHigh |
| ARCN1 | 372 | COPG1 | 22820 | pp | -- | int.I2D: HPRD; int.HPRD: in vivo |
| ARCN1 | 372 | COPA | 1314 | pp | -- | int.I2D: BCI, BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh, HPRD, Krogan_Core; int.HPRD: in vivo |
| ARCN1 | 372 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, Tarassov_PCA, YeastMedium, INTEROLOG |
| COPA | 1314 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid, HPRD, BCI; int.HPRD: in vivo |
| COPA | 1314 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastMedium |
| COPB1 | 1315 | COPG1 | 22820 | pp | -- | int.I2D: BioGrid, HPRD; int.HPRD: in vitro, in vivo, yeast 2-hybrid; int.DIP: MI:0915(physical association) |
| RHOB | 388 | SERTAD1 | 29950 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct, INNATEDB |
| COPB1 | 1315 | SERTAD1 | 29950 | pp | -- | int.Intact: MI:0915(physical association) |
| ARCN1 | 372 | COPB1 | 1315 | pp | -- | int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, BIND, BIND_Yeast, HPRD, Krogan_Core, MIPS; int.HPRD: in vivo |
| RHOB | 388 | PRAF2 | 11230 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: IntAct |
| COPB2 | 9276 | COPG1 | 22820 | pp | -- | int.I2D: BioGrid |
| COPZ1 | 22818 | COPG1 | 22820 | pp | -- | int.I2D: BioGrid, HPRD, BIND; int.HPRD: in vitro, in vivo, yeast 2-hybrid; int.DIP: MI:0915(physical association) |
| ARCN1 | 372 | COPB2 | 9276 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| COPB1 | 1315 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid |
| COPB1 | 1315 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, Tarassov_PCA, YeastMedium; int.DIP: MI:0915(physical association) |
Related GO terms (63)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0030126 | COPI vesicle coat | 6.21e-18 | 8.95e-14 | 9.646 | 6 | 7 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 1.15e-17 | 1.66e-13 | 9.531 | 6 | 8 | 13 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 1.19e-15 | 1.71e-11 | 8.587 | 6 | 8 | 25 |
| GO:0061024 | membrane organization | 7.63e-11 | 1.10e-06 | 6.051 | 6 | 12 | 145 |
| GO:0006886 | intracellular protein transport | 2.08e-10 | 2.99e-06 | 5.813 | 6 | 7 | 171 |
| GO:0006891 | intra-Golgi vesicle-mediated transport | 1.14e-07 | 1.64e-03 | 8.144 | 3 | 4 | 17 |
| GO:0005198 | structural molecule activity | 1.36e-06 | 1.96e-02 | 5.417 | 4 | 6 | 150 |
| GO:0005829 | cytosol | 1.20e-04 | 1.00e+00 | 2.168 | 7 | 61 | 2496 |
| GO:0021691 | cerebellar Purkinje cell layer maturation | 6.24e-04 | 1.00e+00 | 10.646 | 1 | 1 | 1 |
| GO:0051683 | establishment of Golgi localization | 1.87e-03 | 1.00e+00 | 9.061 | 1 | 2 | 3 |
| GO:0072384 | organelle transport along microtubule | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 2 | 4 |
| GO:0010008 | endosome membrane | 3.74e-03 | 1.00e+00 | 4.408 | 2 | 5 | 151 |
| GO:0006927 | transformed cell apoptotic process | 3.74e-03 | 1.00e+00 | 8.061 | 1 | 1 | 6 |
| GO:0030157 | pancreatic juice secretion | 4.36e-03 | 1.00e+00 | 7.839 | 1 | 1 | 7 |
| GO:0005798 | Golgi-associated vesicle | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 10 |
| GO:0015813 | L-glutamate transport | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 10 |
| GO:0030131 | clathrin adaptor complex | 8.70e-03 | 1.00e+00 | 6.839 | 1 | 2 | 14 |
| GO:0043473 | pigmentation | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 20 |
| GO:0071479 | cellular response to ionizing radiation | 1.43e-02 | 1.00e+00 | 6.123 | 1 | 2 | 23 |
| GO:0043231 | intracellular membrane-bounded organelle | 1.57e-02 | 1.00e+00 | 3.333 | 2 | 5 | 318 |
| GO:0015031 | protein transport | 1.80e-02 | 1.00e+00 | 3.233 | 2 | 8 | 341 |
| GO:0008333 | endosome to lysosome transport | 1.80e-02 | 1.00e+00 | 5.788 | 1 | 1 | 29 |
| GO:0000139 | Golgi membrane | 2.00e-02 | 1.00e+00 | 3.150 | 2 | 6 | 361 |
| GO:0032154 | cleavage furrow | 2.35e-02 | 1.00e+00 | 5.398 | 1 | 1 | 38 |
| GO:0045786 | negative regulation of cell cycle | 2.41e-02 | 1.00e+00 | 5.361 | 1 | 1 | 39 |
| GO:0070301 | cellular response to hydrogen peroxide | 2.53e-02 | 1.00e+00 | 5.289 | 1 | 2 | 41 |
| GO:0008344 | adult locomotory behavior | 2.71e-02 | 1.00e+00 | 5.187 | 1 | 1 | 44 |
| GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 2.77e-02 | 1.00e+00 | 5.154 | 1 | 1 | 45 |
| GO:0019003 | GDP binding | 2.90e-02 | 1.00e+00 | 5.092 | 1 | 2 | 47 |
| GO:0007266 | Rho protein signal transduction | 2.90e-02 | 1.00e+00 | 5.092 | 1 | 1 | 47 |
| GO:0000910 | cytokinesis | 3.14e-02 | 1.00e+00 | 4.974 | 1 | 2 | 51 |
| GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment | 3.44e-02 | 1.00e+00 | 4.839 | 1 | 1 | 56 |
| GO:0031902 | late endosome membrane | 4.70e-02 | 1.00e+00 | 4.379 | 1 | 2 | 77 |
| GO:0005179 | hormone activity | 4.82e-02 | 1.00e+00 | 4.342 | 1 | 1 | 79 |
| GO:0005794 | Golgi apparatus | 5.28e-02 | 1.00e+00 | 2.393 | 2 | 9 | 610 |
| GO:0045766 | positive regulation of angiogenesis | 5.36e-02 | 1.00e+00 | 4.187 | 1 | 1 | 88 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 6.95e-02 | 1.00e+00 | 3.801 | 1 | 4 | 115 |
| GO:0016020 | membrane | 7.75e-02 | 1.00e+00 | 1.516 | 3 | 41 | 1681 |
| GO:0001525 | angiogenesis | 1.13e-01 | 1.00e+00 | 3.069 | 1 | 3 | 191 |
| GO:0003924 | GTPase activity | 1.16e-01 | 1.00e+00 | 3.024 | 1 | 7 | 197 |
| GO:0030168 | platelet activation | 1.21e-01 | 1.00e+00 | 2.967 | 1 | 2 | 205 |
| GO:0006184 | GTP catabolic process | 1.26e-01 | 1.00e+00 | 2.905 | 1 | 7 | 214 |
| GO:0043065 | positive regulation of apoptotic process | 1.55e-01 | 1.00e+00 | 2.580 | 1 | 6 | 268 |
| GO:0007264 | small GTPase mediated signal transduction | 1.61e-01 | 1.00e+00 | 2.522 | 1 | 7 | 279 |
| GO:0005525 | GTP binding | 1.78e-01 | 1.00e+00 | 2.370 | 1 | 7 | 310 |
| GO:0007411 | axon guidance | 1.82e-01 | 1.00e+00 | 2.329 | 1 | 3 | 319 |
| GO:0005925 | focal adhesion | 2.07e-01 | 1.00e+00 | 2.130 | 1 | 19 | 366 |
| GO:0007155 | cell adhesion | 2.08e-01 | 1.00e+00 | 2.119 | 1 | 3 | 369 |
| GO:0008284 | positive regulation of cell proliferation | 2.15e-01 | 1.00e+00 | 2.069 | 1 | 7 | 382 |
| GO:0007596 | blood coagulation | 2.51e-01 | 1.00e+00 | 1.816 | 1 | 3 | 455 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 2.62e-01 | 1.00e+00 | 1.745 | 1 | 5 | 478 |
| GO:0016032 | viral process | 2.88e-01 | 1.00e+00 | 1.585 | 1 | 33 | 534 |
| GO:0005783 | endoplasmic reticulum | 2.98e-01 | 1.00e+00 | 1.527 | 1 | 3 | 556 |
| GO:0006915 | apoptotic process | 2.98e-01 | 1.00e+00 | 1.530 | 1 | 9 | 555 |
| GO:0005737 | cytoplasm | 4.31e-01 | 1.00e+00 | 0.352 | 3 | 43 | 3767 |
| GO:0070062 | extracellular vesicular exosome | 4.56e-01 | 1.00e+00 | 0.417 | 2 | 40 | 2400 |
| GO:0005615 | extracellular space | 4.61e-01 | 1.00e+00 | 0.744 | 1 | 2 | 957 |
| GO:0044822 | poly(A) RNA binding | 4.96e-01 | 1.00e+00 | 0.602 | 1 | 32 | 1056 |
| GO:0005886 | plasma membrane | 4.98e-01 | 1.00e+00 | 0.312 | 2 | 20 | 2582 |
| GO:0005515 | protein binding | 5.61e-01 | 1.00e+00 | 0.090 | 4 | 62 | 6024 |
| GO:0006351 | transcription, DNA-templated | 6.14e-01 | 1.00e+00 | 0.148 | 1 | 6 | 1446 |
| GO:0016021 | integral component of membrane | 7.36e-01 | 1.00e+00 | -0.307 | 1 | 5 | 1982 |
| GO:0005634 | nucleus | 9.67e-01 | 1.00e+00 | -1.508 | 1 | 43 | 4559 |