int-snw-2800
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| tai-screen-luciferase | 6.833 | 2.39e-160 | 1.44e-08 | 6.25e-04 |
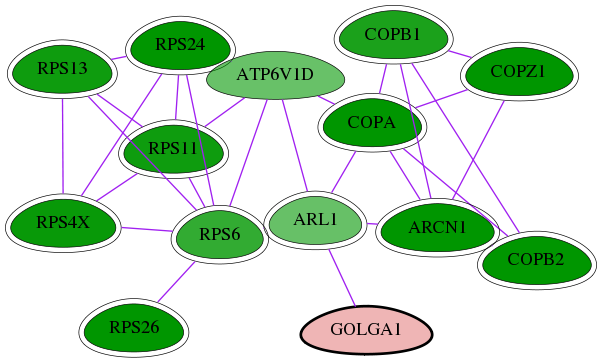
Genes (14)
| Gene Symbol | Entrez Gene ID | Frequency | tai-screen-luciferase gene score | Best subnetwork score | Degree | List-Gonzales GI | Tai-Hits |
|---|---|---|---|---|---|---|---|
| RPS4X | 6191 | 18 | -6.747 | 7.555 | 263 | Yes | - |
| COPZ1 | 22818 | 32 | -8.301 | 9.063 | 13 | Yes | Yes |
| ATP6V1D | 51382 | 8 | -4.131 | 7.286 | 149 | - | - |
| COPA | 1314 | 32 | -9.395 | 9.063 | 170 | Yes | Yes |
| ARL1 | 400 | 4 | -4.142 | 8.046 | 110 | - | Yes |
| RPS11 | 6205 | 17 | -6.588 | 7.555 | 175 | Yes | - |
| RPS6 | 6194 | 17 | -5.603 | 8.046 | 208 | Yes | - |
| RPS24 | 6229 | 21 | -7.034 | 8.389 | 217 | Yes | - |
| RPS26 | 6231 | 17 | -7.478 | 8.046 | 60 | Yes | - |
| ARCN1 | 372 | 32 | -8.232 | 9.063 | 118 | Yes | Yes |
| [ GOLGA1 ] | 2800 | 1 | 2.029 | 6.833 | 5 | - | - |
| COPB1 | 1315 | 22 | -6.221 | 9.063 | 118 | Yes | Yes |
| COPB2 | 9276 | 32 | -13.168 | 9.063 | 41 | Yes | Yes |
| RPS13 | 6207 | 17 | -6.589 | 7.555 | 174 | Yes | - |
Interactions (29)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| RPS11 | 6205 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS4X | 6191 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| ARL1 | 400 | COPA | 1314 | pp | -- | int.I2D: BioGrid_Yeast |
| COPA | 1314 | COPB1 | 1315 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct_Yeast, MINT_Yeast, Krogan_Core, YeastHigh |
| ARCN1 | 372 | COPA | 1314 | pp | -- | int.I2D: BCI, BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh, HPRD, Krogan_Core; int.HPRD: in vivo |
| RPS4X | 6191 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, Tarassov_PCA, YeastMedium, INTEROLOG |
| COPA | 1314 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid, HPRD, BCI; int.HPRD: in vivo |
| RPS11 | 6205 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| COPA | 1314 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastMedium |
| COPA | 1314 | ATP6V1D | 51382 | pp | -- | int.I2D: YeastLow |
| RPS4X | 6191 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast, INTEROLOG, YeastMedium |
| ARCN1 | 372 | ARL1 | 400 | pp | -- | int.I2D: BioGrid_Yeast |
| ARL1 | 400 | RPS6 | 6194 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | COPB1 | 1315 | pp | -- | int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, BIND, BIND_Yeast, HPRD, Krogan_Core, MIPS; int.HPRD: in vivo |
| RPS6 | 6194 | RPS26 | 6231 | pp | -- | int.I2D: YeastLow |
| RPS6 | 6194 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| ARL1 | 400 | GOLGA1 | 2800 | pp | -- | int.I2D: BCI, BioGrid, HPRD; int.HPRD: in vitro, in vivo, yeast 2-hybrid |
| RPS11 | 6205 | RPS13 | 6207 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS11 | 6205 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS6 | 6194 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS4X | 6191 | RPS6 | 6194 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS24 | 6229 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS13 | 6207 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| ARL1 | 400 | ATP6V1D | 51382 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | COPB2 | 9276 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| COPB1 | 1315 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid |
| COPB1 | 1315 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, Tarassov_PCA, YeastMedium; int.DIP: MI:0915(physical association) |
Related GO terms (112)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0030126 | COPI vesicle coat | 3.04e-13 | 4.38e-09 | 8.746 | 5 | 7 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 4.94e-13 | 7.12e-09 | 8.630 | 5 | 8 | 13 |
| GO:0022627 | cytosolic small ribosomal subunit | 7.72e-13 | 1.11e-08 | 7.308 | 6 | 20 | 39 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 2.03e-11 | 2.92e-07 | 7.687 | 5 | 8 | 25 |
| GO:0019083 | viral transcription | 7.53e-11 | 1.09e-06 | 6.254 | 6 | 20 | 81 |
| GO:0006415 | translational termination | 1.17e-10 | 1.68e-06 | 6.151 | 6 | 20 | 87 |
| GO:0006414 | translational elongation | 1.76e-10 | 2.54e-06 | 6.055 | 6 | 20 | 93 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 3.48e-10 | 5.02e-06 | 5.893 | 6 | 20 | 104 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6.09e-10 | 8.78e-06 | 5.761 | 6 | 20 | 114 |
| GO:0019058 | viral life cycle | 6.42e-10 | 9.26e-06 | 5.748 | 6 | 23 | 115 |
| GO:0015935 | small ribosomal subunit | 1.31e-09 | 1.89e-05 | 7.921 | 4 | 9 | 17 |
| GO:0006413 | translational initiation | 1.42e-09 | 2.04e-05 | 5.560 | 6 | 21 | 131 |
| GO:0003735 | structural constituent of ribosome | 1.86e-09 | 2.68e-05 | 5.496 | 6 | 22 | 137 |
| GO:0016071 | mRNA metabolic process | 3.46e-08 | 4.99e-04 | 4.793 | 6 | 25 | 223 |
| GO:0006412 | translation | 4.16e-08 | 6.00e-04 | 4.748 | 6 | 23 | 230 |
| GO:0005829 | cytosol | 4.54e-08 | 6.55e-04 | 2.308 | 12 | 61 | 2496 |
| GO:0016070 | RNA metabolic process | 6.36e-08 | 9.17e-04 | 4.645 | 6 | 25 | 247 |
| GO:0061024 | membrane organization | 1.78e-07 | 2.57e-03 | 5.151 | 5 | 12 | 145 |
| GO:0016032 | viral process | 2.51e-07 | 3.62e-03 | 3.755 | 7 | 33 | 534 |
| GO:0016020 | membrane | 2.90e-07 | 4.18e-03 | 2.616 | 10 | 41 | 1681 |
| GO:0006886 | intracellular protein transport | 4.06e-07 | 5.85e-03 | 4.913 | 5 | 7 | 171 |
| GO:0006891 | intra-Golgi vesicle-mediated transport | 4.91e-07 | 7.08e-03 | 7.506 | 3 | 4 | 17 |
| GO:0044267 | cellular protein metabolic process | 2.93e-06 | 4.22e-02 | 3.705 | 6 | 23 | 474 |
| GO:0033119 | negative regulation of RNA splicing | 8.74e-06 | 1.26e-01 | 8.687 | 2 | 2 | 5 |
| GO:0010467 | gene expression | 2.12e-05 | 3.06e-01 | 3.208 | 6 | 29 | 669 |
| GO:0005840 | ribosome | 2.18e-05 | 3.14e-01 | 5.736 | 3 | 8 | 58 |
| GO:0044822 | poly(A) RNA binding | 2.40e-05 | 3.46e-01 | 2.772 | 7 | 32 | 1056 |
| GO:0042274 | ribosomal small subunit biogenesis | 5.74e-05 | 8.28e-01 | 7.424 | 2 | 6 | 12 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 2.00e-04 | 1.00e+00 | 6.549 | 2 | 3 | 22 |
| GO:0005844 | polysome | 2.19e-04 | 1.00e+00 | 6.485 | 2 | 2 | 23 |
| GO:0019843 | rRNA binding | 3.03e-04 | 1.00e+00 | 6.254 | 2 | 4 | 27 |
| GO:0005198 | structural molecule activity | 3.69e-04 | 1.00e+00 | 4.365 | 3 | 6 | 150 |
| GO:0033176 | proton-transporting V-type ATPase complex | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0021691 | cerebellar Purkinje cell layer maturation | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0022605 | oogenesis stage | 9.71e-04 | 1.00e+00 | 10.009 | 1 | 1 | 1 |
| GO:0003729 | mRNA binding | 2.15e-03 | 1.00e+00 | 4.839 | 2 | 2 | 72 |
| GO:0005794 | Golgi apparatus | 2.26e-03 | 1.00e+00 | 2.756 | 4 | 9 | 610 |
| GO:0009404 | toxin metabolic process | 2.91e-03 | 1.00e+00 | 8.424 | 1 | 1 | 3 |
| GO:0043231 | intracellular membrane-bounded organelle | 3.23e-03 | 1.00e+00 | 3.281 | 3 | 5 | 318 |
| GO:0006364 | rRNA processing | 3.63e-03 | 1.00e+00 | 4.454 | 2 | 7 | 94 |
| GO:0070062 | extracellular vesicular exosome | 4.05e-03 | 1.00e+00 | 1.587 | 7 | 40 | 2400 |
| GO:0005925 | focal adhesion | 4.79e-03 | 1.00e+00 | 3.078 | 3 | 19 | 366 |
| GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 3 | 5 |
| GO:0031584 | activation of phospholipase D activity | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 5 |
| GO:0005802 | trans-Golgi network | 4.94e-03 | 1.00e+00 | 4.227 | 2 | 3 | 110 |
| GO:0030529 | ribonucleoprotein complex | 5.12e-03 | 1.00e+00 | 4.201 | 2 | 7 | 112 |
| GO:0061512 | protein localization to cilium | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 6 |
| GO:0006924 | activation-induced cell death of T cells | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 6 |
| GO:0002309 | T cell proliferation involved in immune response | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 6 |
| GO:0034101 | erythrocyte homeostasis | 6.78e-03 | 1.00e+00 | 7.201 | 1 | 1 | 7 |
| GO:0030157 | pancreatic juice secretion | 6.78e-03 | 1.00e+00 | 7.201 | 1 | 1 | 7 |
| GO:0048193 | Golgi vesicle transport | 6.78e-03 | 1.00e+00 | 7.201 | 1 | 1 | 7 |
| GO:0034067 | protein localization to Golgi apparatus | 6.78e-03 | 1.00e+00 | 7.201 | 1 | 1 | 7 |
| GO:0000028 | ribosomal small subunit assembly | 6.78e-03 | 1.00e+00 | 7.201 | 1 | 3 | 7 |
| GO:0008286 | insulin receptor signaling pathway | 8.11e-03 | 1.00e+00 | 3.859 | 2 | 3 | 142 |
| GO:0005798 | Golgi-associated vesicle | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 10 |
| GO:0000042 | protein targeting to Golgi | 1.06e-02 | 1.00e+00 | 6.549 | 1 | 1 | 11 |
| GO:0031929 | TOR signaling | 1.26e-02 | 1.00e+00 | 6.308 | 1 | 1 | 13 |
| GO:0030131 | clathrin adaptor complex | 1.35e-02 | 1.00e+00 | 6.201 | 1 | 2 | 14 |
| GO:0031369 | translation initiation factor binding | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 15 |
| GO:0048821 | erythrocyte development | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 15 |
| GO:0007369 | gastrulation | 1.83e-02 | 1.00e+00 | 5.761 | 1 | 1 | 19 |
| GO:0043473 | pigmentation | 1.92e-02 | 1.00e+00 | 5.687 | 1 | 1 | 20 |
| GO:0042626 | ATPase activity, coupled to transmembrane movement of substances | 2.50e-02 | 1.00e+00 | 5.308 | 1 | 1 | 26 |
| GO:0015992 | proton transport | 2.78e-02 | 1.00e+00 | 5.151 | 1 | 1 | 29 |
| GO:0033572 | transferrin transport | 2.87e-02 | 1.00e+00 | 5.102 | 1 | 1 | 30 |
| GO:0001890 | placenta development | 2.97e-02 | 1.00e+00 | 5.055 | 1 | 1 | 31 |
| GO:0007093 | mitotic cell cycle checkpoint | 2.97e-02 | 1.00e+00 | 5.055 | 1 | 1 | 31 |
| GO:0051701 | interaction with host | 3.06e-02 | 1.00e+00 | 5.009 | 1 | 1 | 32 |
| GO:0033077 | T cell differentiation in thymus | 3.16e-02 | 1.00e+00 | 4.964 | 1 | 1 | 33 |
| GO:0090382 | phagosome maturation | 3.44e-02 | 1.00e+00 | 4.839 | 1 | 1 | 36 |
| GO:0008047 | enzyme activator activity | 3.72e-02 | 1.00e+00 | 4.723 | 1 | 1 | 39 |
| GO:0042147 | retrograde transport, endosome to Golgi | 3.91e-02 | 1.00e+00 | 4.651 | 1 | 1 | 41 |
| GO:0045727 | positive regulation of translation | 4.19e-02 | 1.00e+00 | 4.549 | 1 | 1 | 44 |
| GO:0008344 | adult locomotory behavior | 4.19e-02 | 1.00e+00 | 4.549 | 1 | 1 | 44 |
| GO:0032580 | Golgi cisterna membrane | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 1 | 45 |
| GO:0044297 | cell body | 4.38e-02 | 1.00e+00 | 4.485 | 1 | 1 | 46 |
| GO:0007030 | Golgi organization | 4.65e-02 | 1.00e+00 | 4.394 | 1 | 2 | 49 |
| GO:0000139 | Golgi membrane | 4.66e-02 | 1.00e+00 | 2.513 | 2 | 6 | 361 |
| GO:0005737 | cytoplasm | 4.77e-02 | 1.00e+00 | 0.937 | 7 | 43 | 3767 |
| GO:0006879 | cellular iron ion homeostasis | 4.84e-02 | 1.00e+00 | 4.336 | 1 | 1 | 51 |
| GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment | 5.30e-02 | 1.00e+00 | 4.201 | 1 | 1 | 56 |
| GO:0005730 | nucleolus | 6.58e-02 | 1.00e+00 | 1.328 | 4 | 24 | 1641 |
| GO:0005929 | cilium | 7.22e-02 | 1.00e+00 | 3.742 | 1 | 1 | 77 |
| GO:0005179 | hormone activity | 7.40e-02 | 1.00e+00 | 3.705 | 1 | 1 | 79 |
| GO:0042384 | cilium assembly | 8.04e-02 | 1.00e+00 | 3.582 | 1 | 1 | 86 |
| GO:0042593 | glucose homeostasis | 8.13e-02 | 1.00e+00 | 3.566 | 1 | 1 | 87 |
| GO:0048471 | perinuclear region of cytoplasm | 8.35e-02 | 1.00e+00 | 2.037 | 2 | 7 | 502 |
| GO:0001649 | osteoblast differentiation | 8.57e-02 | 1.00e+00 | 3.485 | 1 | 3 | 92 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.35e-01 | 1.00e+00 | 2.790 | 1 | 9 | 149 |
| GO:0019904 | protein domain specific binding | 1.60e-01 | 1.00e+00 | 2.525 | 1 | 3 | 179 |
| GO:0003924 | GTPase activity | 1.75e-01 | 1.00e+00 | 2.387 | 1 | 7 | 197 |
| GO:0005765 | lysosomal membrane | 1.80e-01 | 1.00e+00 | 2.343 | 1 | 2 | 203 |
| GO:0006184 | GTP catabolic process | 1.89e-01 | 1.00e+00 | 2.267 | 1 | 7 | 214 |
| GO:0007067 | mitotic nuclear division | 1.99e-01 | 1.00e+00 | 2.182 | 1 | 5 | 227 |
| GO:0030425 | dendrite | 2.02e-01 | 1.00e+00 | 2.163 | 1 | 2 | 230 |
| GO:0000166 | nucleotide binding | 2.24e-01 | 1.00e+00 | 1.992 | 1 | 2 | 259 |
| GO:0043065 | positive regulation of apoptotic process | 2.31e-01 | 1.00e+00 | 1.943 | 1 | 6 | 268 |
| GO:0007264 | small GTPase mediated signal transduction | 2.39e-01 | 1.00e+00 | 1.885 | 1 | 7 | 279 |
| GO:0005525 | GTP binding | 2.62e-01 | 1.00e+00 | 1.733 | 1 | 7 | 310 |
| GO:0019901 | protein kinase binding | 2.67e-01 | 1.00e+00 | 1.700 | 1 | 6 | 317 |
| GO:0007275 | multicellular organismal development | 2.67e-01 | 1.00e+00 | 1.705 | 1 | 1 | 316 |
| GO:0005813 | centrosome | 2.74e-01 | 1.00e+00 | 1.660 | 1 | 5 | 326 |
| GO:0008284 | positive regulation of cell proliferation | 3.13e-01 | 1.00e+00 | 1.431 | 1 | 7 | 382 |
| GO:0043066 | negative regulation of apoptotic process | 3.42e-01 | 1.00e+00 | 1.281 | 1 | 13 | 424 |
| GO:0055085 | transmembrane transport | 3.49e-01 | 1.00e+00 | 1.244 | 1 | 3 | 435 |
| GO:0005783 | endoplasmic reticulum | 4.23e-01 | 1.00e+00 | 0.890 | 1 | 3 | 556 |
| GO:0005515 | protein binding | 5.68e-01 | 1.00e+00 | 0.037 | 6 | 62 | 6024 |
| GO:0005615 | extracellular space | 6.18e-01 | 1.00e+00 | 0.106 | 1 | 2 | 957 |
| GO:0046872 | metal ion binding | 7.36e-01 | 1.00e+00 | -0.343 | 1 | 10 | 1307 |
| GO:0005634 | nucleus | 8.68e-01 | 1.00e+00 | -0.561 | 3 | 43 | 4559 |
| GO:0005886 | plasma membrane | 9.37e-01 | 1.00e+00 | -1.326 | 1 | 20 | 2582 |