int-snw-23433
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| tai-screen-luciferase | 6.337 | 2.49e-134 | 4.62e-07 | 2.89e-03 |
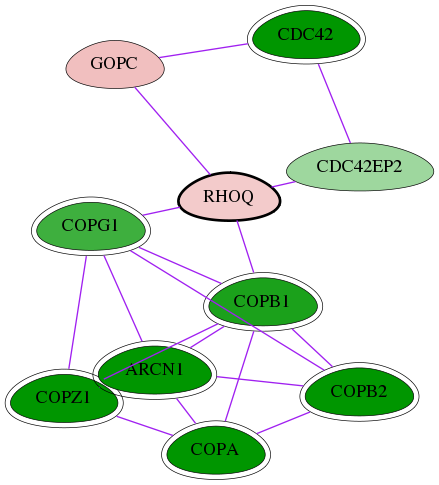
Genes (10)
| Gene Symbol | Entrez Gene ID | Frequency | tai-screen-luciferase gene score | Best subnetwork score | Degree | List-Gonzales GI | Tai-Hits |
|---|---|---|---|---|---|---|---|
| COPZ1 | 22818 | 32 | -8.301 | 9.063 | 13 | Yes | Yes |
| [ RHOQ ] | 23433 | 1 | 1.414 | 6.337 | 19 | - | - |
| COPA | 1314 | 32 | -9.395 | 9.063 | 170 | Yes | Yes |
| CDC42EP2 | 10435 | 1 | -2.649 | 6.337 | 5 | - | - |
| CDC42 | 998 | 11 | -6.960 | 8.389 | 265 | Yes | Yes |
| GOPC | 57120 | 1 | 1.749 | 6.337 | 31 | - | - |
| ARCN1 | 372 | 32 | -8.232 | 9.063 | 118 | Yes | Yes |
| COPB2 | 9276 | 32 | -13.168 | 9.063 | 41 | Yes | Yes |
| COPG1 | 22820 | 9 | -5.279 | 7.138 | 37 | Yes | Yes |
| COPB1 | 1315 | 22 | -6.221 | 9.063 | 118 | Yes | Yes |
Interactions (19)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| CDC42 | 998 | CDC42EP2 | 10435 | pp | -- | int.Intact: MI:0915(physical association); int.I2D: BioGrid, HPRD, BIND, IntAct, SOURAV_MAPK_HIGH; int.HPRD: in vitro, in vivo |
| ARCN1 | 372 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, Tarassov_PCA, YeastMedium, INTEROLOG |
| COPA | 1314 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid, HPRD, BCI; int.HPRD: in vivo |
| COPG1 | 22820 | RHOQ | 23433 | pp | -- | int.I2D: HPRD; int.HPRD: in vivo |
| ARCN1 | 372 | COPB1 | 1315 | pp | -- | int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, BIND, BIND_Yeast, HPRD, Krogan_Core, MIPS; int.HPRD: in vivo |
| ARCN1 | 372 | COPB2 | 9276 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| COPB1 | 1315 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, Tarassov_PCA, YeastMedium; int.DIP: MI:0915(physical association) |
| CDC42 | 998 | GOPC | 57120 | pp | -- | int.I2D: BioGrid |
| COPA | 1314 | COPB1 | 1315 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct_Yeast, MINT_Yeast, Krogan_Core, YeastHigh |
| ARCN1 | 372 | COPG1 | 22820 | pp | -- | int.I2D: HPRD; int.HPRD: in vivo |
| ARCN1 | 372 | COPA | 1314 | pp | -- | int.I2D: BCI, BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh, HPRD, Krogan_Core; int.HPRD: in vivo |
| COPA | 1314 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastMedium |
| COPB1 | 1315 | RHOQ | 23433 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| COPB1 | 1315 | COPG1 | 22820 | pp | -- | int.I2D: BioGrid, HPRD; int.HPRD: in vitro, in vivo, yeast 2-hybrid; int.DIP: MI:0915(physical association) |
| CDC42EP2 | 10435 | RHOQ | 23433 | pp | -- | int.I2D: BioGrid, HPRD, BIND; int.HPRD: in vitro, yeast 2-hybrid |
| COPB2 | 9276 | COPG1 | 22820 | pp | -- | int.I2D: BioGrid |
| COPZ1 | 22818 | COPG1 | 22820 | pp | -- | int.I2D: BioGrid, HPRD, BIND; int.HPRD: in vitro, in vivo, yeast 2-hybrid; int.DIP: MI:0915(physical association) |
| COPB1 | 1315 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid |
| RHOQ | 23433 | GOPC | 57120 | pp | -- | int.I2D: BioGrid, HPRD; int.HPRD: in vitro, in vivo, yeast 2-hybrid |
Related GO terms (149)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0030126 | COPI vesicle coat | 1.55e-17 | 2.24e-13 | 9.494 | 6 | 7 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 2.88e-17 | 4.15e-13 | 9.379 | 6 | 8 | 13 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 2.96e-15 | 4.28e-11 | 8.435 | 6 | 8 | 25 |
| GO:0061024 | membrane organization | 1.89e-10 | 2.73e-06 | 5.899 | 6 | 12 | 145 |
| GO:0006886 | intracellular protein transport | 5.14e-10 | 7.41e-06 | 5.661 | 6 | 7 | 171 |
| GO:0006891 | intra-Golgi vesicle-mediated transport | 1.62e-07 | 2.34e-03 | 7.992 | 3 | 4 | 17 |
| GO:0051683 | establishment of Golgi localization | 1.30e-06 | 1.87e-02 | 9.909 | 2 | 2 | 3 |
| GO:0005198 | structural molecule activity | 2.25e-06 | 3.24e-02 | 5.265 | 4 | 6 | 150 |
| GO:0072384 | organelle transport along microtubule | 2.59e-06 | 3.74e-02 | 9.494 | 2 | 2 | 4 |
| GO:0031274 | positive regulation of pseudopodium assembly | 1.94e-05 | 2.80e-01 | 8.172 | 2 | 2 | 10 |
| GO:0005829 | cytosol | 2.57e-05 | 3.71e-01 | 2.209 | 8 | 61 | 2496 |
| GO:0000139 | Golgi membrane | 7.19e-05 | 1.00e+00 | 3.998 | 4 | 6 | 361 |
| GO:0016020 | membrane | 3.44e-04 | 1.00e+00 | 2.364 | 6 | 41 | 1681 |
| GO:0032427 | GBD domain binding | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0021691 | cerebellar Purkinje cell layer maturation | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0043004 | cytoplasmic sequestering of CFTR protein | 6.93e-04 | 1.00e+00 | 10.494 | 1 | 1 | 1 |
| GO:0071338 | positive regulation of hair follicle cell proliferation | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0090135 | actin filament branching | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0060661 | submandibular salivary gland formation | 1.39e-03 | 1.00e+00 | 9.494 | 1 | 1 | 2 |
| GO:0034191 | apolipoprotein A-I receptor binding | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0003161 | cardiac conduction system development | 2.08e-03 | 1.00e+00 | 8.909 | 1 | 1 | 3 |
| GO:0008360 | regulation of cell shape | 2.19e-03 | 1.00e+00 | 4.808 | 2 | 2 | 103 |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 2.72e-03 | 1.00e+00 | 4.649 | 2 | 4 | 115 |
| GO:0060684 | epithelial-mesenchymal cell signaling | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0051835 | positive regulation of synapse structural plasticity | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0048664 | neuron fate determination | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0001515 | opioid peptide activity | 2.77e-03 | 1.00e+00 | 8.494 | 1 | 1 | 4 |
| GO:0030036 | actin cytoskeleton organization | 3.10e-03 | 1.00e+00 | 4.552 | 2 | 2 | 123 |
| GO:0035088 | establishment or maintenance of apical/basal cell polarity | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0036336 | dendritic cell migration | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0031256 | leading edge membrane | 3.46e-03 | 1.00e+00 | 8.172 | 1 | 1 | 5 |
| GO:0060789 | hair follicle placode formation | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0007097 | nuclear migration | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0003334 | keratinocyte development | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0048554 | positive regulation of metalloenzyme activity | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0043497 | regulation of protein heterodimerization activity | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 6 |
| GO:0030157 | pancreatic juice secretion | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0090005 | negative regulation of establishment of protein localization to plasma membrane | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0006893 | Golgi to plasma membrane transport | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0051489 | regulation of filopodium assembly | 4.84e-03 | 1.00e+00 | 7.687 | 1 | 1 | 7 |
| GO:0007289 | spermatid nucleus differentiation | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0005522 | profilin binding | 5.53e-03 | 1.00e+00 | 7.494 | 1 | 1 | 8 |
| GO:0090136 | epithelial cell-cell adhesion | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0017049 | GTP-Rho binding | 6.22e-03 | 1.00e+00 | 7.324 | 1 | 1 | 9 |
| GO:0005798 | Golgi-associated vesicle | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 1 | 10 |
| GO:0060047 | heart contraction | 6.91e-03 | 1.00e+00 | 7.172 | 1 | 1 | 10 |
| GO:0005794 | Golgi apparatus | 7.23e-03 | 1.00e+00 | 2.826 | 3 | 9 | 610 |
| GO:0045176 | apical protein localization | 7.60e-03 | 1.00e+00 | 7.035 | 1 | 1 | 11 |
| GO:0003924 | GTPase activity | 7.77e-03 | 1.00e+00 | 3.872 | 2 | 7 | 197 |
| GO:0030140 | trans-Golgi network transport vesicle | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 12 |
| GO:0031334 | positive regulation of protein complex assembly | 8.98e-03 | 1.00e+00 | 6.794 | 1 | 1 | 13 |
| GO:0006184 | GTP catabolic process | 9.12e-03 | 1.00e+00 | 3.753 | 2 | 7 | 214 |
| GO:0031333 | negative regulation of protein complex assembly | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 14 |
| GO:0031996 | thioesterase binding | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 1 | 14 |
| GO:0030131 | clathrin adaptor complex | 9.67e-03 | 1.00e+00 | 6.687 | 1 | 2 | 14 |
| GO:0051233 | spindle midzone | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 2 | 15 |
| GO:0030225 | macrophage differentiation | 1.04e-02 | 1.00e+00 | 6.587 | 1 | 1 | 15 |
| GO:0042176 | regulation of protein catabolic process | 1.10e-02 | 1.00e+00 | 6.494 | 1 | 3 | 16 |
| GO:0030742 | GTP-dependent protein binding | 1.17e-02 | 1.00e+00 | 6.407 | 1 | 1 | 17 |
| GO:0090316 | positive regulation of intracellular protein transport | 1.17e-02 | 1.00e+00 | 6.407 | 1 | 1 | 17 |
| GO:0007088 | regulation of mitosis | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 18 |
| GO:0046847 | filopodium assembly | 1.31e-02 | 1.00e+00 | 6.246 | 1 | 1 | 19 |
| GO:0030866 | cortical actin cytoskeleton organization | 1.31e-02 | 1.00e+00 | 6.246 | 1 | 1 | 19 |
| GO:0043473 | pigmentation | 1.38e-02 | 1.00e+00 | 6.172 | 1 | 1 | 20 |
| GO:0051491 | positive regulation of filopodium assembly | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 21 |
| GO:0031435 | mitogen-activated protein kinase kinase kinase binding | 1.45e-02 | 1.00e+00 | 6.102 | 1 | 1 | 21 |
| GO:0007264 | small GTPase mediated signal transduction | 1.51e-02 | 1.00e+00 | 3.370 | 2 | 7 | 279 |
| GO:0031424 | keratinization | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 1 | 22 |
| GO:0036464 | cytoplasmic ribonucleoprotein granule | 1.52e-02 | 1.00e+00 | 6.035 | 1 | 3 | 22 |
| GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 1 | 23 |
| GO:0005100 | Rho GTPase activator activity | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 1 | 23 |
| GO:0002040 | sprouting angiogenesis | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 1 | 23 |
| GO:0051017 | actin filament bundle assembly | 1.58e-02 | 1.00e+00 | 5.971 | 1 | 1 | 23 |
| GO:0007163 | establishment or maintenance of cell polarity | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 24 |
| GO:0046326 | positive regulation of glucose import | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 24 |
| GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 2 | 26 |
| GO:0045859 | regulation of protein kinase activity | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 26 |
| GO:0005083 | small GTPase regulator activity | 1.79e-02 | 1.00e+00 | 5.794 | 1 | 1 | 26 |
| GO:0005525 | GTP binding | 1.85e-02 | 1.00e+00 | 3.218 | 2 | 7 | 310 |
| GO:0051149 | positive regulation of muscle cell differentiation | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 1 | 27 |
| GO:0032467 | positive regulation of cytokinesis | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 1 | 27 |
| GO:0031069 | hair follicle morphogenesis | 1.86e-02 | 1.00e+00 | 5.739 | 1 | 1 | 27 |
| GO:0043231 | intracellular membrane-bounded organelle | 1.94e-02 | 1.00e+00 | 3.181 | 2 | 5 | 318 |
| GO:0072686 | mitotic spindle | 1.99e-02 | 1.00e+00 | 5.636 | 1 | 1 | 29 |
| GO:0032956 | regulation of actin cytoskeleton organization | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 2 | 30 |
| GO:0031647 | regulation of protein stability | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 1 | 30 |
| GO:0005886 | plasma membrane | 2.08e-02 | 1.00e+00 | 1.482 | 5 | 20 | 2582 |
| GO:0030838 | positive regulation of actin filament polymerization | 2.33e-02 | 1.00e+00 | 5.407 | 1 | 1 | 34 |
| GO:0042692 | muscle cell differentiation | 2.33e-02 | 1.00e+00 | 5.407 | 1 | 1 | 34 |
| GO:0005737 | cytoplasm | 2.44e-02 | 1.00e+00 | 1.200 | 6 | 43 | 3767 |
| GO:0034332 | adherens junction organization | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 2 | 36 |
| GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 3 | 36 |
| GO:0045740 | positive regulation of DNA replication | 2.47e-02 | 1.00e+00 | 5.324 | 1 | 1 | 36 |
| GO:0007015 | actin filament organization | 2.54e-02 | 1.00e+00 | 5.285 | 1 | 1 | 37 |
| GO:0021762 | substantia nigra development | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 1 | 44 |
| GO:0034613 | cellular protein localization | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 2 | 44 |
| GO:0008344 | adult locomotory behavior | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 1 | 44 |
| GO:0005884 | actin filament | 3.01e-02 | 1.00e+00 | 5.035 | 1 | 1 | 44 |
| GO:0043525 | positive regulation of neuron apoptotic process | 3.14e-02 | 1.00e+00 | 4.971 | 1 | 2 | 46 |
| GO:0007030 | Golgi organization | 3.35e-02 | 1.00e+00 | 4.879 | 1 | 2 | 49 |
| GO:0030175 | filopodium | 3.41e-02 | 1.00e+00 | 4.850 | 1 | 2 | 50 |
| GO:0006888 | ER to Golgi vesicle-mediated transport | 3.62e-02 | 1.00e+00 | 4.766 | 1 | 1 | 53 |
| GO:0046330 | positive regulation of JNK cascade | 3.75e-02 | 1.00e+00 | 4.713 | 1 | 1 | 55 |
| GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment | 3.82e-02 | 1.00e+00 | 4.687 | 1 | 1 | 56 |
| GO:0012505 | endomembrane system | 3.82e-02 | 1.00e+00 | 4.687 | 1 | 1 | 56 |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 4.02e-02 | 1.00e+00 | 4.612 | 1 | 1 | 59 |
| GO:0030141 | secretory granule | 4.22e-02 | 1.00e+00 | 4.540 | 1 | 1 | 62 |
| GO:0032869 | cellular response to insulin stimulus | 4.28e-02 | 1.00e+00 | 4.517 | 1 | 2 | 63 |
| GO:0031295 | T cell costimulation | 4.42e-02 | 1.00e+00 | 4.472 | 1 | 3 | 65 |
| GO:0035264 | multicellular organism growth | 4.55e-02 | 1.00e+00 | 4.428 | 1 | 1 | 67 |
| GO:0050790 | regulation of catalytic activity | 4.68e-02 | 1.00e+00 | 4.386 | 1 | 1 | 69 |
| GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 5.02e-02 | 1.00e+00 | 4.285 | 1 | 2 | 74 |
| GO:0044325 | ion channel binding | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 3 | 75 |
| GO:0060070 | canonical Wnt signaling pathway | 5.08e-02 | 1.00e+00 | 4.265 | 1 | 1 | 75 |
| GO:0010629 | negative regulation of gene expression | 5.21e-02 | 1.00e+00 | 4.227 | 1 | 2 | 77 |
| GO:0005179 | hormone activity | 5.35e-02 | 1.00e+00 | 4.190 | 1 | 1 | 79 |
| GO:0045177 | apical part of cell | 5.48e-02 | 1.00e+00 | 4.154 | 1 | 1 | 81 |
| GO:0032321 | positive regulation of Rho GTPase activity | 5.54e-02 | 1.00e+00 | 4.137 | 1 | 1 | 82 |
| GO:0014069 | postsynaptic density | 7.05e-02 | 1.00e+00 | 3.780 | 1 | 3 | 105 |
| GO:0030496 | midbody | 7.24e-02 | 1.00e+00 | 3.739 | 1 | 2 | 108 |
| GO:0005815 | microtubule organizing center | 7.31e-02 | 1.00e+00 | 3.726 | 1 | 3 | 109 |
| GO:0015630 | microtubule cytoskeleton | 7.37e-02 | 1.00e+00 | 3.713 | 1 | 3 | 110 |
| GO:0005911 | cell-cell junction | 9.04e-02 | 1.00e+00 | 3.407 | 1 | 2 | 136 |
| GO:0008286 | insulin receptor signaling pathway | 9.42e-02 | 1.00e+00 | 3.344 | 1 | 3 | 142 |
| GO:0051260 | protein homooligomerization | 9.61e-02 | 1.00e+00 | 3.314 | 1 | 2 | 145 |
| GO:0010628 | positive regulation of gene expression | 9.68e-02 | 1.00e+00 | 3.304 | 1 | 4 | 146 |
| GO:0045121 | membrane raft | 1.03e-01 | 1.00e+00 | 3.209 | 1 | 3 | 156 |
| GO:0043005 | neuron projection | 1.04e-01 | 1.00e+00 | 3.200 | 1 | 6 | 157 |
| GO:0045211 | postsynaptic membrane | 1.12e-01 | 1.00e+00 | 3.085 | 1 | 4 | 170 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 1.24e-01 | 1.00e+00 | 2.932 | 1 | 4 | 189 |
| GO:0030425 | dendrite | 1.49e-01 | 1.00e+00 | 2.649 | 1 | 2 | 230 |
| GO:0043025 | neuronal cell body | 1.57e-01 | 1.00e+00 | 2.558 | 1 | 4 | 245 |
| GO:0043234 | protein complex | 1.87e-01 | 1.00e+00 | 2.290 | 1 | 6 | 295 |
| GO:0019901 | protein kinase binding | 1.99e-01 | 1.00e+00 | 2.186 | 1 | 6 | 317 |
| GO:0007411 | axon guidance | 2.00e-01 | 1.00e+00 | 2.177 | 1 | 3 | 319 |
| GO:0030054 | cell junction | 2.10e-01 | 1.00e+00 | 2.102 | 1 | 4 | 336 |
| GO:0015031 | protein transport | 2.13e-01 | 1.00e+00 | 2.081 | 1 | 8 | 341 |
| GO:0070062 | extracellular vesicular exosome | 2.24e-01 | 1.00e+00 | 0.850 | 3 | 40 | 2400 |
| GO:0005925 | focal adhesion | 2.27e-01 | 1.00e+00 | 1.978 | 1 | 19 | 366 |
| GO:0007596 | blood coagulation | 2.74e-01 | 1.00e+00 | 1.664 | 1 | 3 | 455 |
| GO:0042802 | identical protein binding | 2.89e-01 | 1.00e+00 | 1.575 | 1 | 3 | 484 |
| GO:0016032 | viral process | 3.14e-01 | 1.00e+00 | 1.433 | 1 | 33 | 534 |
| GO:0005783 | endoplasmic reticulum | 3.25e-01 | 1.00e+00 | 1.375 | 1 | 3 | 556 |
| GO:0045087 | innate immune response | 3.44e-01 | 1.00e+00 | 1.275 | 1 | 8 | 596 |
| GO:0005515 | protein binding | 4.12e-01 | 1.00e+00 | 0.260 | 5 | 62 | 6024 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 4.30e-01 | 1.00e+00 | 0.870 | 1 | 6 | 789 |
| GO:0005615 | extracellular space | 4.97e-01 | 1.00e+00 | 0.592 | 1 | 2 | 957 |
| GO:0044822 | poly(A) RNA binding | 5.33e-01 | 1.00e+00 | 0.450 | 1 | 32 | 1056 |