int-snw-23193
Statistics
| Dataset | Score | p-value 1 | p-value 2 | p-value 3 |
|---|---|---|---|---|
| tai-screen-luciferase | 6.468 | 5.52e-141 | 1.92e-07 | 1.97e-03 |
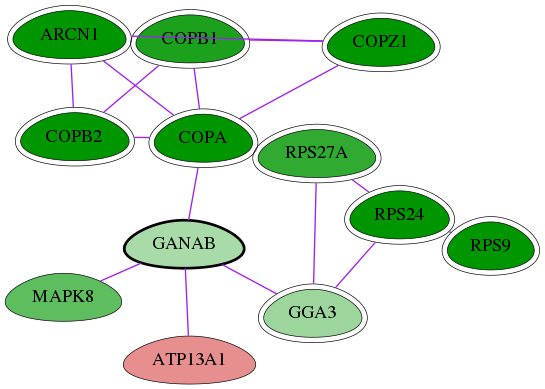
Genes (12)
| Gene Symbol | Entrez Gene ID | Frequency | tai-screen-luciferase gene score | Best subnetwork score | Degree | List-Gonzales GI | Tai-Hits |
|---|---|---|---|---|---|---|---|
| MAPK8 | 5599 | 2 | -4.403 | 6.468 | 153 | - | - |
| COPZ1 | 22818 | 32 | -8.301 | 9.063 | 13 | Yes | Yes |
| ATP13A1 | 57130 | 1 | 3.062 | 6.468 | 110 | - | - |
| COPA | 1314 | 32 | -9.395 | 9.063 | 170 | Yes | Yes |
| RPS27A | 6233 | 15 | -5.631 | 8.389 | 342 | Yes | - |
| RPS24 | 6229 | 21 | -7.034 | 8.389 | 217 | Yes | - |
| GGA3 | 23163 | 1 | -2.668 | 6.468 | 62 | Yes | - |
| [ GANAB ] | 23193 | 1 | -2.374 | 6.468 | 83 | - | - |
| ARCN1 | 372 | 32 | -8.232 | 9.063 | 118 | Yes | Yes |
| RPS9 | 6203 | 18 | -7.127 | 7.555 | 140 | Yes | - |
| COPB2 | 9276 | 32 | -13.168 | 9.063 | 41 | Yes | Yes |
| COPB1 | 1315 | 22 | -6.221 | 9.063 | 118 | Yes | Yes |
Interactions (18)
| Gene Symbol 1 | Entrez Gene ID 1 | Gene Symbol 2 | Entrez Gene ID 2 | Type | Direction | Origin databases / Sources |
|---|---|---|---|---|---|---|
| RPS9 | 6203 | RPS24 | 6229 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS27A | 6233 | GGA3 | 23163 | pp | -- | int.I2D: BCI, BioGrid_Yeast, BIND, HPRD; int.HPRD: in vitro, yeast 2-hybrid |
| COPA | 1314 | COPB1 | 1315 | pp | -- | int.I2D: BioGrid, BioGrid_Yeast, BIND_Yeast, IntAct_Yeast, MINT_Yeast, Krogan_Core, YeastHigh |
| MAPK8 | 5599 | GANAB | 23193 | pp | -- | int.Intact: MI:0915(physical association) |
| GANAB | 23193 | ATP13A1 | 57130 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | COPB1 | 1315 | pp | -- | int.I2D: BCI, BioGrid, BioGrid_Yeast, IntAct_Yeast, INTEROLOG, MINT_Yeast, YeastHigh, BIND, BIND_Yeast, HPRD, Krogan_Core, MIPS; int.HPRD: in vivo |
| COPA | 1314 | RPS27A | 6233 | pp | -- | int.I2D: YeastLow |
| ARCN1 | 372 | COPA | 1314 | pp | -- | int.I2D: BCI, BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, YeastHigh, HPRD, Krogan_Core; int.HPRD: in vivo |
| COPA | 1314 | GANAB | 23193 | pp | -- | int.I2D: IntAct_Yeast |
| ARCN1 | 372 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, MINT_Yeast, Tarassov_PCA, YeastMedium, INTEROLOG |
| COPA | 1314 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid, HPRD, BCI; int.HPRD: in vivo |
| GGA3 | 23163 | GANAB | 23193 | pp | -- | int.I2D: BioGrid_Yeast |
| ARCN1 | 372 | COPB2 | 9276 | pp | -- | int.I2D: HPRD, BCI; int.HPRD: in vivo |
| COPA | 1314 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, YeastMedium |
| COPB1 | 1315 | COPB2 | 9276 | pp | -- | int.I2D: BioGrid |
| RPS24 | 6229 | GGA3 | 23163 | pp | -- | int.I2D: BioGrid_Yeast |
| RPS24 | 6229 | RPS27A | 6233 | pp | -- | int.I2D: YeastMedium |
| COPB1 | 1315 | COPZ1 | 22818 | pp | -- | int.I2D: BioGrid_Yeast, IntAct_Yeast, Krogan_Core, MINT_Yeast, Tarassov_PCA, YeastMedium; int.DIP: MI:0915(physical association) |
Related GO terms (176)
| Accession number | Name | Hypergeometric test | Corrected p-value | Enrichment ratio | Occurrence in subnetwork | Occurrences in all snw genes | Occurrences in all int/reg genes |
|---|---|---|---|---|---|---|---|
| GO:0030126 | COPI vesicle coat | 1.20e-13 | 1.74e-09 | 8.968 | 5 | 7 | 12 |
| GO:0048205 | COPI coating of Golgi vesicle | 1.95e-13 | 2.82e-09 | 8.853 | 5 | 8 | 13 |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 8.03e-12 | 1.16e-07 | 7.909 | 5 | 8 | 25 |
| GO:0061024 | membrane organization | 8.18e-10 | 1.18e-05 | 5.636 | 6 | 12 | 145 |
| GO:0006886 | intracellular protein transport | 2.22e-09 | 3.20e-05 | 5.398 | 6 | 7 | 171 |
| GO:0006891 | intra-Golgi vesicle-mediated transport | 2.97e-07 | 4.29e-03 | 7.729 | 3 | 4 | 17 |
| GO:0022627 | cytosolic small ribosomal subunit | 3.95e-06 | 5.70e-02 | 6.531 | 3 | 20 | 39 |
| GO:0016020 | membrane | 1.08e-05 | 1.55e-01 | 2.516 | 8 | 41 | 1681 |
| GO:0005829 | cytosol | 1.83e-05 | 2.63e-01 | 2.116 | 9 | 61 | 2496 |
| GO:0019083 | viral transcription | 3.62e-05 | 5.22e-01 | 5.476 | 3 | 20 | 81 |
| GO:0006415 | translational termination | 4.48e-05 | 6.47e-01 | 5.373 | 3 | 20 | 87 |
| GO:0006414 | translational elongation | 5.47e-05 | 7.90e-01 | 5.277 | 3 | 20 | 93 |
| GO:0030131 | clathrin adaptor complex | 5.74e-05 | 8.28e-01 | 7.424 | 2 | 2 | 14 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 7.64e-05 | 1.00e+00 | 5.116 | 3 | 20 | 104 |
| GO:0015935 | small ribosomal subunit | 8.57e-05 | 1.00e+00 | 7.144 | 2 | 9 | 17 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1.00e-04 | 1.00e+00 | 4.983 | 3 | 20 | 114 |
| GO:0019058 | viral life cycle | 1.03e-04 | 1.00e+00 | 4.971 | 3 | 23 | 115 |
| GO:0006413 | translational initiation | 1.52e-04 | 1.00e+00 | 4.783 | 3 | 21 | 131 |
| GO:0003735 | structural constituent of ribosome | 1.73e-04 | 1.00e+00 | 4.718 | 3 | 22 | 137 |
| GO:0005198 | structural molecule activity | 2.26e-04 | 1.00e+00 | 4.587 | 3 | 6 | 150 |
| GO:0044267 | cellular protein metabolic process | 4.62e-04 | 1.00e+00 | 3.342 | 4 | 23 | 474 |
| GO:0016071 | mRNA metabolic process | 7.24e-04 | 1.00e+00 | 4.015 | 3 | 25 | 223 |
| GO:0016032 | viral process | 7.25e-04 | 1.00e+00 | 3.170 | 4 | 33 | 534 |
| GO:0007254 | JNK cascade | 7.30e-04 | 1.00e+00 | 5.616 | 2 | 3 | 49 |
| GO:0006412 | translation | 7.92e-04 | 1.00e+00 | 3.971 | 3 | 23 | 230 |
| GO:0021691 | cerebellar Purkinje cell layer maturation | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 1 |
| GO:0033919 | glucan 1,3-alpha-glucosidase activity | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 1 |
| GO:0017177 | glucosidase II complex | 8.32e-04 | 1.00e+00 | 10.231 | 1 | 1 | 1 |
| GO:0051403 | stress-activated MAPK cascade | 8.87e-04 | 1.00e+00 | 5.476 | 2 | 3 | 54 |
| GO:0016070 | RNA metabolic process | 9.74e-04 | 1.00e+00 | 3.868 | 3 | 25 | 247 |
| GO:0044822 | poly(A) RNA binding | 1.07e-03 | 1.00e+00 | 2.509 | 5 | 32 | 1056 |
| GO:0034146 | toll-like receptor 5 signaling pathway | 1.28e-03 | 1.00e+00 | 5.209 | 2 | 4 | 65 |
| GO:0034166 | toll-like receptor 10 signaling pathway | 1.28e-03 | 1.00e+00 | 5.209 | 2 | 4 | 65 |
| GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway | 1.53e-03 | 1.00e+00 | 5.081 | 2 | 4 | 71 |
| GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway | 1.53e-03 | 1.00e+00 | 5.081 | 2 | 4 | 71 |
| GO:0034162 | toll-like receptor 9 signaling pathway | 1.57e-03 | 1.00e+00 | 5.061 | 2 | 4 | 72 |
| GO:0034134 | toll-like receptor 2 signaling pathway | 1.61e-03 | 1.00e+00 | 5.041 | 2 | 4 | 73 |
| GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 1.75e-03 | 1.00e+00 | 4.983 | 2 | 4 | 76 |
| GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 1.84e-03 | 1.00e+00 | 4.946 | 2 | 4 | 78 |
| GO:0034138 | toll-like receptor 3 signaling pathway | 1.89e-03 | 1.00e+00 | 4.927 | 2 | 4 | 79 |
| GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 1.93e-03 | 1.00e+00 | 4.909 | 2 | 4 | 80 |
| GO:0090045 | positive regulation of deacetylase activity | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 3 |
| GO:0035033 | histone deacetylase regulator activity | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 3 |
| GO:2000017 | positive regulation of determination of dorsal identity | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 3 |
| GO:0004705 | JUN kinase activity | 2.49e-03 | 1.00e+00 | 8.646 | 1 | 1 | 3 |
| GO:0034142 | toll-like receptor 4 signaling pathway | 2.77e-03 | 1.00e+00 | 4.646 | 2 | 4 | 96 |
| GO:0007258 | JUN phosphorylation | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 1 | 4 |
| GO:0031063 | regulation of histone deacetylation | 3.32e-03 | 1.00e+00 | 8.231 | 1 | 1 | 4 |
| GO:0002224 | toll-like receptor signaling pathway | 3.55e-03 | 1.00e+00 | 4.463 | 2 | 4 | 109 |
| GO:0097190 | apoptotic signaling pathway | 3.81e-03 | 1.00e+00 | 4.411 | 2 | 3 | 113 |
| GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 3 | 5 |
| GO:0097300 | programmed necrotic cell death | 4.15e-03 | 1.00e+00 | 7.909 | 1 | 1 | 5 |
| GO:0030306 | ADP-ribosylation factor binding | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 1 | 6 |
| GO:0045182 | translation regulator activity | 4.98e-03 | 1.00e+00 | 7.646 | 1 | 2 | 6 |
| GO:0030157 | pancreatic juice secretion | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 7 |
| GO:0034101 | erythrocyte homeostasis | 5.81e-03 | 1.00e+00 | 7.424 | 1 | 1 | 7 |
| GO:0010008 | endosome membrane | 6.71e-03 | 1.00e+00 | 3.993 | 2 | 5 | 151 |
| GO:0019829 | cation-transporting ATPase activity | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 1 | 9 |
| GO:0071732 | cellular response to nitric oxide | 7.47e-03 | 1.00e+00 | 7.061 | 1 | 1 | 9 |
| GO:0038095 | Fc-epsilon receptor signaling pathway | 8.24e-03 | 1.00e+00 | 3.839 | 2 | 3 | 168 |
| GO:0005798 | Golgi-associated vesicle | 8.29e-03 | 1.00e+00 | 6.909 | 1 | 1 | 10 |
| GO:0019082 | viral protein processing | 9.94e-03 | 1.00e+00 | 6.646 | 1 | 2 | 12 |
| GO:0042274 | ribosomal small subunit biogenesis | 9.94e-03 | 1.00e+00 | 6.646 | 1 | 6 | 12 |
| GO:0032479 | regulation of type I interferon production | 1.08e-02 | 1.00e+00 | 6.531 | 1 | 2 | 13 |
| GO:0031369 | translation initiation factor binding | 1.24e-02 | 1.00e+00 | 6.324 | 1 | 1 | 15 |
| GO:0005794 | Golgi apparatus | 1.24e-02 | 1.00e+00 | 2.563 | 3 | 9 | 610 |
| GO:0075733 | intracellular transport of virus | 1.41e-02 | 1.00e+00 | 6.144 | 1 | 3 | 17 |
| GO:0019068 | virion assembly | 1.41e-02 | 1.00e+00 | 6.144 | 1 | 2 | 17 |
| GO:0010467 | gene expression | 1.60e-02 | 1.00e+00 | 2.430 | 3 | 29 | 669 |
| GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 20 |
| GO:0043473 | pigmentation | 1.65e-02 | 1.00e+00 | 5.909 | 1 | 1 | 20 |
| GO:0007220 | Notch receptor processing | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 2 | 22 |
| GO:0046686 | response to cadmium ion | 1.82e-02 | 1.00e+00 | 5.772 | 1 | 1 | 22 |
| GO:0043065 | positive regulation of apoptotic process | 2.01e-02 | 1.00e+00 | 3.165 | 2 | 6 | 268 |
| GO:0048011 | neurotrophin TRK receptor signaling pathway | 2.02e-02 | 1.00e+00 | 3.160 | 2 | 4 | 269 |
| GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 2.06e-02 | 1.00e+00 | 5.587 | 1 | 2 | 25 |
| GO:0005978 | glycogen biosynthetic process | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 2 | 26 |
| GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 1 | 26 |
| GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2.14e-02 | 1.00e+00 | 5.531 | 1 | 2 | 26 |
| GO:0019843 | rRNA binding | 2.22e-02 | 1.00e+00 | 5.476 | 1 | 4 | 27 |
| GO:0032091 | negative regulation of protein binding | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 1 | 32 |
| GO:0032480 | negative regulation of type I interferon production | 2.63e-02 | 1.00e+00 | 5.231 | 1 | 2 | 32 |
| GO:0043231 | intracellular membrane-bounded organelle | 2.76e-02 | 1.00e+00 | 2.918 | 2 | 5 | 318 |
| GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 2.96e-02 | 1.00e+00 | 5.061 | 1 | 3 | 36 |
| GO:0018107 | peptidyl-threonine phosphorylation | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 1 | 37 |
| GO:0032880 | regulation of protein localization | 3.04e-02 | 1.00e+00 | 5.022 | 1 | 1 | 37 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 3.36e-02 | 1.00e+00 | 4.874 | 1 | 3 | 41 |
| GO:0070301 | cellular response to hydrogen peroxide | 3.36e-02 | 1.00e+00 | 4.874 | 1 | 2 | 41 |
| GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 3.52e-02 | 1.00e+00 | 4.805 | 1 | 2 | 43 |
| GO:0008344 | adult locomotory behavior | 3.60e-02 | 1.00e+00 | 4.772 | 1 | 1 | 44 |
| GO:0009411 | response to UV | 3.60e-02 | 1.00e+00 | 4.772 | 1 | 1 | 44 |
| GO:0006950 | response to stress | 3.76e-02 | 1.00e+00 | 4.708 | 1 | 1 | 46 |
| GO:0098655 | cation transmembrane transport | 3.92e-02 | 1.00e+00 | 4.646 | 1 | 1 | 48 |
| GO:0030666 | endocytic vesicle membrane | 4.32e-02 | 1.00e+00 | 4.503 | 1 | 2 | 53 |
| GO:0097193 | intrinsic apoptotic signaling pathway | 4.48e-02 | 1.00e+00 | 4.450 | 1 | 1 | 55 |
| GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment | 4.56e-02 | 1.00e+00 | 4.424 | 1 | 1 | 56 |
| GO:0016197 | endosomal transport | 4.64e-02 | 1.00e+00 | 4.398 | 1 | 3 | 57 |
| GO:0043066 | negative regulation of apoptotic process | 4.68e-02 | 1.00e+00 | 2.503 | 2 | 13 | 424 |
| GO:0005840 | ribosome | 4.72e-02 | 1.00e+00 | 4.373 | 1 | 8 | 58 |
| GO:0032481 | positive regulation of type I interferon production | 4.96e-02 | 1.00e+00 | 4.300 | 1 | 3 | 61 |
| GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 4.96e-02 | 1.00e+00 | 4.300 | 1 | 2 | 61 |
| GO:0006417 | regulation of translation | 5.04e-02 | 1.00e+00 | 4.277 | 1 | 2 | 62 |
| GO:0001503 | ossification | 5.20e-02 | 1.00e+00 | 4.231 | 1 | 1 | 64 |
| GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 5.20e-02 | 1.00e+00 | 4.231 | 1 | 6 | 64 |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 6 | 65 |
| GO:0071260 | cellular response to mechanical stimulus | 5.28e-02 | 1.00e+00 | 4.209 | 1 | 2 | 65 |
| GO:0018105 | peptidyl-serine phosphorylation | 5.59e-02 | 1.00e+00 | 4.123 | 1 | 4 | 69 |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.67e-02 | 1.00e+00 | 4.102 | 1 | 8 | 70 |
| GO:0042826 | histone deacetylase binding | 5.83e-02 | 1.00e+00 | 4.061 | 1 | 1 | 72 |
| GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 5.83e-02 | 1.00e+00 | 4.061 | 1 | 6 | 72 |
| GO:0051439 | regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 5.99e-02 | 1.00e+00 | 4.022 | 1 | 8 | 74 |
| GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 6.07e-02 | 1.00e+00 | 4.002 | 1 | 6 | 75 |
| GO:0005179 | hormone activity | 6.38e-02 | 1.00e+00 | 3.927 | 1 | 1 | 79 |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 6.38e-02 | 1.00e+00 | 3.927 | 1 | 8 | 79 |
| GO:0018279 | protein N-linked glycosylation via asparagine | 6.54e-02 | 1.00e+00 | 3.891 | 1 | 1 | 81 |
| GO:0071222 | cellular response to lipopolysaccharide | 6.54e-02 | 1.00e+00 | 3.891 | 1 | 3 | 81 |
| GO:0050852 | T cell receptor signaling pathway | 6.85e-02 | 1.00e+00 | 3.822 | 1 | 2 | 85 |
| GO:0000187 | activation of MAPK activity | 7.24e-02 | 1.00e+00 | 3.739 | 1 | 2 | 90 |
| GO:0042470 | melanosome | 7.32e-02 | 1.00e+00 | 3.723 | 1 | 2 | 91 |
| GO:0071456 | cellular response to hypoxia | 7.39e-02 | 1.00e+00 | 3.708 | 1 | 3 | 92 |
| GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 7.39e-02 | 1.00e+00 | 3.708 | 1 | 6 | 92 |
| GO:0006364 | rRNA processing | 7.55e-02 | 1.00e+00 | 3.677 | 1 | 7 | 94 |
| GO:0006915 | apoptotic process | 7.56e-02 | 1.00e+00 | 2.115 | 2 | 9 | 555 |
| GO:0045087 | innate immune response | 8.56e-02 | 1.00e+00 | 2.012 | 2 | 8 | 596 |
| GO:0005802 | trans-Golgi network | 8.78e-02 | 1.00e+00 | 3.450 | 1 | 3 | 110 |
| GO:0030529 | ribonucleoprotein complex | 8.93e-02 | 1.00e+00 | 3.424 | 1 | 7 | 112 |
| GO:0000209 | protein polyubiquitination | 9.24e-02 | 1.00e+00 | 3.373 | 1 | 7 | 116 |
| GO:0016192 | vesicle-mediated transport | 9.32e-02 | 1.00e+00 | 3.361 | 1 | 1 | 117 |
| GO:0006006 | glucose metabolic process | 9.39e-02 | 1.00e+00 | 3.348 | 1 | 3 | 118 |
| GO:0007219 | Notch signaling pathway | 9.54e-02 | 1.00e+00 | 3.324 | 1 | 3 | 120 |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 9.85e-02 | 1.00e+00 | 3.277 | 1 | 3 | 124 |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | 1.01e-01 | 1.00e+00 | 3.231 | 1 | 3 | 128 |
| GO:0030246 | carbohydrate binding | 1.06e-01 | 1.00e+00 | 3.165 | 1 | 1 | 134 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 1.08e-01 | 1.00e+00 | 3.144 | 1 | 5 | 136 |
| GO:0006457 | protein folding | 1.13e-01 | 1.00e+00 | 3.071 | 1 | 2 | 143 |
| GO:0005788 | endoplasmic reticulum lumen | 1.14e-01 | 1.00e+00 | 3.051 | 1 | 1 | 145 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 1.14e-01 | 1.00e+00 | 3.051 | 1 | 3 | 145 |
| GO:0010628 | positive regulation of gene expression | 1.15e-01 | 1.00e+00 | 3.041 | 1 | 4 | 146 |
| GO:0034220 | ion transmembrane transport | 1.17e-01 | 1.00e+00 | 3.012 | 1 | 2 | 149 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 1.17e-01 | 1.00e+00 | 3.012 | 1 | 9 | 149 |
| GO:0042981 | regulation of apoptotic process | 1.18e-01 | 1.00e+00 | 3.002 | 1 | 7 | 150 |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | 1.22e-01 | 1.00e+00 | 2.946 | 1 | 3 | 156 |
| GO:0070062 | extracellular vesicular exosome | 1.25e-01 | 1.00e+00 | 1.002 | 4 | 40 | 2400 |
| GO:0043687 | post-translational protein modification | 1.27e-01 | 1.00e+00 | 2.891 | 1 | 1 | 162 |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | 1.43e-01 | 1.00e+00 | 2.708 | 1 | 3 | 184 |
| GO:0007173 | epidermal growth factor receptor signaling pathway | 1.46e-01 | 1.00e+00 | 2.669 | 1 | 4 | 189 |
| GO:0019221 | cytokine-mediated signaling pathway | 1.69e-01 | 1.00e+00 | 2.443 | 1 | 2 | 221 |
| GO:0005737 | cytoplasm | 1.81e-01 | 1.00e+00 | 0.674 | 5 | 43 | 3767 |
| GO:0005975 | carbohydrate metabolic process | 1.91e-01 | 1.00e+00 | 2.248 | 1 | 3 | 253 |
| GO:0000166 | nucleotide binding | 1.95e-01 | 1.00e+00 | 2.214 | 1 | 2 | 259 |
| GO:0006281 | DNA repair | 1.97e-01 | 1.00e+00 | 2.203 | 1 | 4 | 261 |
| GO:0006200 | ATP catabolic process | 2.18e-01 | 1.00e+00 | 2.041 | 1 | 3 | 292 |
| GO:0005654 | nucleoplasm | 2.26e-01 | 1.00e+00 | 1.152 | 2 | 15 | 1082 |
| GO:0004674 | protein serine/threonine kinase activity | 2.28e-01 | 1.00e+00 | 1.964 | 1 | 5 | 308 |
| GO:0000139 | Golgi membrane | 2.62e-01 | 1.00e+00 | 1.735 | 1 | 6 | 361 |
| GO:0005925 | focal adhesion | 2.65e-01 | 1.00e+00 | 1.715 | 1 | 19 | 366 |
| GO:0008284 | positive regulation of cell proliferation | 2.75e-01 | 1.00e+00 | 1.654 | 1 | 7 | 382 |
| GO:0008150 | biological_process | 2.80e-01 | 1.00e+00 | 1.627 | 1 | 3 | 389 |
| GO:0000278 | mitotic cell cycle | 2.81e-01 | 1.00e+00 | 1.620 | 1 | 11 | 391 |
| GO:0005524 | ATP binding | 2.95e-01 | 1.00e+00 | 0.889 | 2 | 15 | 1298 |
| GO:0046872 | metal ion binding | 2.98e-01 | 1.00e+00 | 0.879 | 2 | 10 | 1307 |
| GO:0055085 | transmembrane transport | 3.08e-01 | 1.00e+00 | 1.466 | 1 | 3 | 435 |
| GO:0003674 | molecular_function | 3.13e-01 | 1.00e+00 | 1.437 | 1 | 1 | 444 |
| GO:0005783 | endoplasmic reticulum | 3.76e-01 | 1.00e+00 | 1.112 | 1 | 3 | 556 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 3.84e-01 | 1.00e+00 | 1.076 | 1 | 7 | 570 |
| GO:0005789 | endoplasmic reticulum membrane | 3.84e-01 | 1.00e+00 | 1.076 | 1 | 3 | 570 |
| GO:0005730 | nucleolus | 4.04e-01 | 1.00e+00 | 0.551 | 2 | 24 | 1641 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 4.91e-01 | 1.00e+00 | 0.607 | 1 | 6 | 789 |
| GO:0005615 | extracellular space | 5.61e-01 | 1.00e+00 | 0.329 | 1 | 2 | 957 |
| GO:0005739 | mitochondrion | 5.77e-01 | 1.00e+00 | 0.268 | 1 | 8 | 998 |
| GO:0044281 | small molecule metabolic process | 6.51e-01 | 1.00e+00 | -0.011 | 1 | 15 | 1211 |
| GO:0005886 | plasma membrane | 6.61e-01 | 1.00e+00 | -0.103 | 2 | 20 | 2582 |
| GO:0006351 | transcription, DNA-templated | 7.19e-01 | 1.00e+00 | -0.267 | 1 | 6 | 1446 |
| GO:0005634 | nucleus | 7.84e-01 | 1.00e+00 | -0.338 | 3 | 43 | 4559 |
| GO:0005515 | protein binding | 8.10e-01 | 1.00e+00 | -0.325 | 4 | 62 | 6024 |
| GO:0016021 | integral component of membrane | 8.30e-01 | 1.00e+00 | -0.722 | 1 | 5 | 1982 |